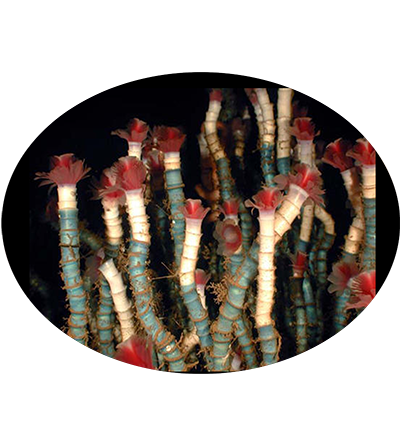
Lamellibrachia luymesi is a species of tube worms in the family Siboglinidae. It lives at deep-sea cold seeps where hydrocarbons (oil and methane) are leaking out of the seafloor. It is entirely reliant on internal, sulfide-oxidizing bacterial symbionts for its nutrition. These are located in a centrally located 'trophosome'. It provides the bacteria with hydrogen sulfide and oxygen by taking them up from the environment and binding them to a specialized hemoglobin molecule. It may help fuel the generation of sulfide by excreting sulfate through their roots into the sediments below the aggregations.
| Species | Phylum | Common Name | Ecosystem | Depth | Habitat | NCBI Taxonomy ID |
|---|---|---|---|---|---|---|
| Lamellibrachia luymesi | Annelida | cold seep-dwelling tubeworm Lamellibrachia luymesi | Hydrothermal vent | >540 | the Gulf of Mexico (N 28° 11.58′, W 89° 47.94′) | 238240 |
| Genome Assembly | Genome Size | Assembly level | Released year | WGS accession | Submitter | BioProject | BUSCO completeness | Gene Number |
|---|---|---|---|---|---|---|---|---|
| LLUY_1.0 | ~688Mb | Scaffold | 2019 | SDWI01 | Auburn University | PRJNA516467 | ~95% | 38,998 |
| Title | Journal | Pubmed ID |
|---|---|---|
| Genomic adaptations to chemosymbiosis in the deep-sea seep-dwelling tubeworm Lamellibrachia luymesi | BMC Biology | 31739792 |
| Gene ID | Description |
|---|---|
| LLU|FUN_014003-T1 | |
| LLU|FUN_014004-T1 | SODIUM/CHLORIDE DEPENDENT TRANSPORTER |
| LLU|FUN_014005-T1 | SODIUM/CHLORIDE DEPENDENT TRANSPORTER |
| LLU|FUN_014006-T1 | |
| LLU|FUN_014007-T1 | |
| LLU|FUN_014008-T1 | |
| LLU|FUN_014009-T1 | PARP/ZINC FINGER CCCH TYPE DOMAIN CONTAINING PROTEIN |
| LLU|FUN_014010-T1 | G-PROTEIN COUPLED RECEPTOR |
| LLU|FUN_014011-T1 | GUANYLATE CYCLASE SOLUBLE SUBUNIT BETA-2 |
| LLU|FUN_014012-T1 | AMIDOPHOSPHORIBOSYLTRANSFERASE |
| LLU|FUN_014013-T1 | ZGC:153352 |
| LLU|FUN_014014-T1 | |
| LLU|FUN_014015-T1 | PLAT DOMAIN-CONTAINING PROTEIN |
| LLU|FUN_014016-T1 | |
| LLU|FUN_014017-T1 | |
| LLU|FUN_014018-T1 | EPHRIN TYPE-B RECEPTOR |
| LLU|FUN_014019-T1 | |
| LLU|FUN_014020-T1 | POLYCYSTIN FAMILY MEMBER |
| LLU|FUN_014021-T1 | POLYCYSTIN FAMILY MEMBER |
| LLU|FUN_014022-T1 | POLYCYSTIN-1 |
| LLU|FUN_014023-T1 | PULMONARY SURFACTANT-ASSOCIATED PROTEIN A |
| LLU|FUN_014024-T1 | BTB/POZ DOMAIN-CONTAINING ADAPTER FOR CUL3-MEDIATED RHOA DEGRADATION PROTEIN FAMILY MEMBER |
| LLU|FUN_014025-T1 | |
| LLU|FUN_014026-T1 | |
| LLU|FUN_014027-T1 | |
| LLU|FUN_014028-T1 | |
| LLU|FUN_014029-T1 | |
| LLU|FUN_014030-T1 | SERINE PROTEASE INHIBITOR, SERPIN |
| LLU|FUN_014037-T1 | HOMEOBOX PROTEIN HHEX |
| LLU|FUN_014038-T1 | MANNOSE, PHOSPHOLIPASE, LECTIN RECEPTOR RELATED |
| LLU|FUN_014039-T1 | |
| LLU|FUN_014040-T1 | |
| LLU|FUN_014041-T1 | |
| LLU|FUN_014042-T1 | BLR2807 PROTEIN |
| LLU|FUN_014043-T1 | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE DEAMINASE-RELATED |
| LLU|FUN_014044-T1 | |
| LLU|FUN_014045-T1 | |
| LLU|FUN_014046-T1 | |
| LLU|FUN_014047-T1 | |
| LLU|FUN_014048-T1 | |
| LLU|FUN_014049-T1 | |
| LLU|FUN_014050-T1 | METALLOPROTEASE M41 FTSH |
| LLU|FUN_014051-T1 | CARBOXYLASE:PYRUVATE/ACETYL-COA/PROPIONYL-COA CARBOXYLASE |
| LLU|FUN_014052-T1 | GLUTAMATE 5-KINASE |
| LLU|FUN_014053-T1 | MAP-KINASE ACTIVATING DEATH DOMAIN PROTEIN MADD /DENN/AEX-3 C.ELEGANS |
| LLU|FUN_014054-T1 | DUAL SPECIFICITY PROTEIN KINASE |
| LLU|FUN_014055-T1 | MUCIN-17 |
| LLU|FUN_014056-T1 | DUAL SPECIFICITY PROTEIN KINASE |
| LLU|FUN_014057-T1 | |
| LLU|FUN_014058-T1 | SLIT RELATED LEUCINE-RICH REPEAT NEURONAL PROTEIN |

