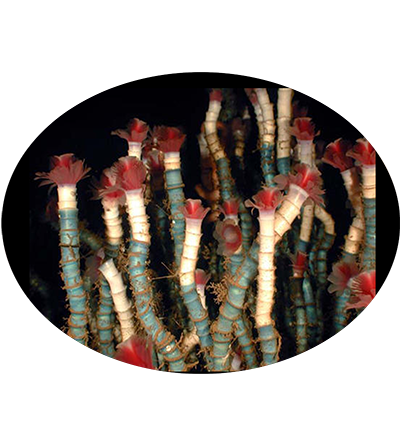
Lamellibrachia luymesi is a species of tube worms in the family Siboglinidae. It lives at deep-sea cold seeps where hydrocarbons (oil and methane) are leaking out of the seafloor. It is entirely reliant on internal, sulfide-oxidizing bacterial symbionts for its nutrition. These are located in a centrally located 'trophosome'. It provides the bacteria with hydrogen sulfide and oxygen by taking them up from the environment and binding them to a specialized hemoglobin molecule. It may help fuel the generation of sulfide by excreting sulfate through their roots into the sediments below the aggregations.
| Species | Phylum | Common Name | Ecosystem | Depth | Habitat | NCBI Taxonomy ID |
|---|---|---|---|---|---|---|
| Lamellibrachia luymesi | Annelida | cold seep-dwelling tubeworm Lamellibrachia luymesi | Hydrothermal vent | >540 | the Gulf of Mexico (N 28° 11.58′, W 89° 47.94′) | 238240 |
| Genome Assembly | Genome Size | Assembly level | Released year | WGS accession | Submitter | BioProject | BUSCO completeness | Gene Number |
|---|---|---|---|---|---|---|---|---|
| LLUY_1.0 | ~688Mb | Scaffold | 2019 | SDWI01 | Auburn University | PRJNA516467 | ~95% | 38,998 |
| Title | Journal | Pubmed ID |
|---|---|---|
| Genomic adaptations to chemosymbiosis in the deep-sea seep-dwelling tubeworm Lamellibrachia luymesi | BMC Biology | 31739792 |
| Gene ID | Description |
|---|---|
| LLU|FUN_020523-T1 | CASEIN KINASE-RELATED |
| LLU|FUN_020524-T1 | TRNA-DIHYDROURIDINE(20) SYNTHASE [NAD(P)+]-LIKE |
| LLU|FUN_020525-T1 | PAR-6-RELATED |
| LLU|FUN_020526-T1 | SELECTIN LIGAND RELATED |
| LLU|FUN_020527-T1 | SELECTIN LIGAND RELATED |
| LLU|FUN_020528-T1 | CYSTEINE AND HISTIDINE-RICH PROTEIN 1 |
| LLU|FUN_020529-T1 | PHOSPHATIDYLINOSITIDE PHOSPHATASE SAC1 |
| LLU|FUN_020530-T1 | ENGULFMENT AND CELL MOTILITY |
| LLU|FUN_020531-T1 | PIEZO-TYPE MECHANOSENSITIVE ION CHANNEL HOMOLOG |
| LLU|FUN_020532-T1 | |
| LLU|FUN_020533-T1 | |
| LLU|FUN_020534-T1 | |
| LLU|FUN_020535-T1 | NEUROTRANSMITTER GATED ION CHANNEL |
| LLU|FUN_020536-T1 | |
| LLU|FUN_020537-T1 | PARP/ZINC FINGER CCCH TYPE DOMAIN CONTAINING PROTEIN |
| LLU|FUN_020538-T1 | MAP/MICROTUBULE AFFINITY-REGULATING KINASE |
| LLU|FUN_020539-T1 | |
| LLU|FUN_020540-T1 | |
| LLU|FUN_020541-T1 | CARTILAGE INTERMEDIATE LAYER PROTEIN CLIP |
| LLU|FUN_020542-T1 | |
| LLU|FUN_020543-T1 | ANKYRIN REPEAT FAMILY PROTEIN |
| LLU|FUN_020544-T1 | |
| LLU|FUN_020545-T1 | SRCR DOMAIN-CONTAINING PROTEIN |
| LLU|FUN_020546-T1 | |
| LLU|FUN_020547-T1 | PLECKSTRIN HOMOLOGY DOMAIN-CONTAINING FAMILY G MEMBER 7 |
| LLU|FUN_020548-T1 | |
| LLU|FUN_020549-T1 | PLECKSTRIN HOMOLOGY DOMAIN-CONTAINING FAMILY G MEMBER 7 |
| LLU|FUN_020550-T1 | |
| LLU|FUN_020551-T1 | |
| LLU|FUN_020552-T1 | |
| LLU|FUN_020553-T1 | |
| LLU|FUN_020554-T1 | |
| LLU|FUN_020555-T1 | CHLORIDE CHANNEL PROTEIN 2 |
| LLU|FUN_020556-T1 | |
| LLU|FUN_020557-T1 | |
| LLU|FUN_020558-T1 | CHLORIDE CHANNEL PROTEIN 2 |
| LLU|FUN_020559-T1 | |
| LLU|FUN_020560-T1 | D-2-HYDROXYGLUTARATE DEHYDROGENASE, MITOCHONDRIAL |
| LLU|FUN_020561-T1 | GLYCOSYLTRANSFERASE 14 FAMILY MEMBER |
| LLU|FUN_020562-T1 | DNA REPLICATION-RELATED ELEMENT FACTOR, ISOFORM A |
| LLU|FUN_020563-T1 | |
| LLU|FUN_020564-T1 | TRANSLOCON-ASSOCIATED PROTEIN, ALPHA SUBUNIT |
| LLU|FUN_020565-T1 | |
| LLU|FUN_020566-T1 | |
| LLU|FUN_020567-T1 | |
| LLU|FUN_020568-T1 | |
| LLU|FUN_020569-T1 | |
| LLU|FUN_020570-T1 | ATAXIN 1 |
| LLU|FUN_020571-T1 | ATAXIN 1 |
| LLU|FUN_020572-T1 |

