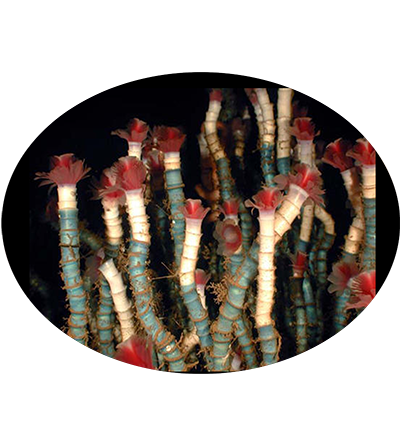
Lamellibrachia luymesi is a species of tube worms in the family Siboglinidae. It lives at deep-sea cold seeps where hydrocarbons (oil and methane) are leaking out of the seafloor. It is entirely reliant on internal, sulfide-oxidizing bacterial symbionts for its nutrition. These are located in a centrally located 'trophosome'. It provides the bacteria with hydrogen sulfide and oxygen by taking them up from the environment and binding them to a specialized hemoglobin molecule. It may help fuel the generation of sulfide by excreting sulfate through their roots into the sediments below the aggregations.
Animalia (Kingdom); Annelida (Phylum); Polychaeta (Class); Sedentaria (Subclass); Canalipalpata (Infraclass); Sabellida (Order); Siboglinidae (Family); Lamellibrachia (Genus); Lamellibrachia luymesi (Species)
Lamellibrachia luymesi van der Land & Nørrevang, 1975
1. VAN DER LAND J. The systematic position of Lamellibrachia (Annelida, Vestimentifera)[J]. Zeitschrift fur Zoologische Systematik und Evolutionsforschung, Sonderheft, 1975, 1975: 86-101. (Land et al., 1975)
Not stated. Perhaps (from the locality information given) named after a research vessel named "Luymes", in turn named after the hydrographer Johan Lambertus Hendrikus Luymes
continental slope off Guyana, S America, North Atlantic Ocean, 8.0167, -57.4° (8 1' N, 57 24' W) "Luymes" Guyana Shelf Exp. sta. 101, depth 500 m
| Species | Phylum | Common Name | Ecosystem | Depth | Habitat | NCBI Taxonomy ID |
|---|---|---|---|---|---|---|
| Lamellibrachia luymesi | Annelida | cold seep-dwelling tubeworm Lamellibrachia luymesi | Hydrothermal vent | >540 | the Gulf of Mexico (N 28° 11.58′, W 89° 47.94′) | 238240 |
| Genome Assembly | Genome Size | Assembly level | Released year | WGS accession | Submitter | BioProject | BUSCO completeness (%) | Scaffold/Contig N50 (kb) | GC content (%) | Repeat Rate (%) | Gene Number |
|---|---|---|---|---|---|---|---|---|---|---|---|
| LLUY_1.0 | ~688Mb | Scaffold | 2019 | SDWI01 | Auburn University | PRJNA516467 | 95.80 | 373/24.1 | 40.16 | 36.92 | 38,998 |
| Title | Journal | Pubmed ID |
|---|---|---|
| Genomic adaptations to chemosymbiosis in the deep-sea seep-dwelling tubeworm Lamellibrachia luymesi | BMC Biology | 31739792 |
| Gene ID | Description |
|---|---|
| LLU|FUN_000021-T1 | - |
| LLU|FUN_000022-T1 | - |
| LLU|FUN_000023-T1 | - |
| LLU|FUN_000024-T1 | 40S RIBOSOMAL PROTEIN S12 |
| LLU|FUN_000025-T1 | - |
| LLU|FUN_000026-T1 | - |
| LLU|FUN_000027-T1 | NOTCH LIGAND FAMILY MEMBER |
| LLU|FUN_000028-T1 | - |
| LLU|FUN_000029-T1 | EGF-LIKE DOMAIN-CONTAINING PROTEIN |
| LLU|FUN_000030-T1 | - |
| LLU|FUN_000031-T1 | - |
| LLU|FUN_000032-T1 | - |
| LLU|FUN_000033-T1 | - |
| LLU|FUN_000034-T1 | - |
| LLU|FUN_000035-T1 | - |
| LLU|FUN_000036-T1 | - |
| LLU|FUN_000037-T1 | BASIC HELIX-LOOP-HELIX TRANSCRIPTION FACTOR, TWIST |
| LLU|FUN_000038-T1 | - |
| LLU|FUN_000039-T1 | BASIC HELIX-LOOP-HELIX TRANSCRIPTION FACTOR, TWIST |
| LLU|FUN_000040-T1 | STRUCTURAL MAINTENANCE OF CHROMOSOMES 5,6 SMC5, SMC6 |
| Annotation ID | Description | Type | Subtype | GeneRatio | Evolution Type | P-value | Q-value |
|---|---|---|---|---|---|---|---|
| PTHR44025 | LOW-DENSITY LIPOPROTEIN RECEPTOR-RELATED PROTEIN | PANTHER | - | - | expanded | - | - |
| PTHR12011 | G-PROTEIN COUPLED RECEPTOR | PANTHER | - | - | expanded | - | - |
| PTHR11177 | CHITINASE | PANTHER | - | - | expanded | - | - |
| PTHR28576 | PIGGYBAC TRANSPOSABLE ELEMENT-DERIVED PROTEIN | PANTHER | - | - | expanded | - | - |
| PTHR19325 | COMPLEMENT COMPONENT-RELATED SUSHI DOMAIN-CONTAINING | PANTHER | - | - | expanded | - | - |
| PTHR19325 | COMPLEMENT COMPONENT-RELATED SUSHI DOMAIN-CONTAINING | PANTHER | - | - | expanded | - | - |
| PTHR11119 | XANTHINE-URACIL / VITAMIN C PERMEASE FAMILY MEMBER | PANTHER | - | - | expanded | - | - |
| PTHR10877 | POLYCYSTIN-RELATED | PANTHER | - | - | expanded | - | - |
| PTHR13802 | MUCIN 4-RELATED | PANTHER | - | - | expanded | - | - |
| PTHR10131 | TNF RECEPTOR ASSOCIATED FACTOR | PANTHER | - | - | expanded | - | - |
| PTHR11908 | XANTHINE DEHYDROGENASE | PANTHER | - | - | expanded | - | - |
| PTHR11709 | MULTI-COPPER OXIDASE | PANTHER | - | - | expanded | - | - |
| PTHR14647 | GALACTOSE-3-O-SULFOTRANSFERASE | PANTHER | - | - | expanded | - | - |
| PTHR23033 | BETA1,3-GALACTOSYLTRANSFERASE | PANTHER | - | - | expanded | - | - |
| PTHR10283 | SOLUTE CARRIER FAMILY 13 MEMBER | PANTHER | - | - | expanded | - | - |
| PTHR24039 | FIBRILLIN | PANTHER | - | - | expanded | - | - |
| PTHR14453 | PARP/ZINC FINGER CCCH TYPE DOMAIN CONTAINING PROTEIN | PANTHER | - | - | expanded | - | - |
| PTHR14453 | PARP/ZINC FINGER CCCH TYPE DOMAIN CONTAINING PROTEIN | PANTHER | - | - | expanded | - | - |
| PTHR24221 | ABC TRANSPORTER | PANTHER | - | - | expanded | - | - |
| PTHR24033 | FAMILY NOT NAMED | PANTHER | - | - | expanded | - | - |
| PTHR10579 | CALCIUM-ACTIVATED CHLORIDE CHANNEL REGULATOR | PANTHER | - | - | expanded | - | - |
| PTHR11039 | NEBULIN | PANTHER | - | - | expanded | - | - |

