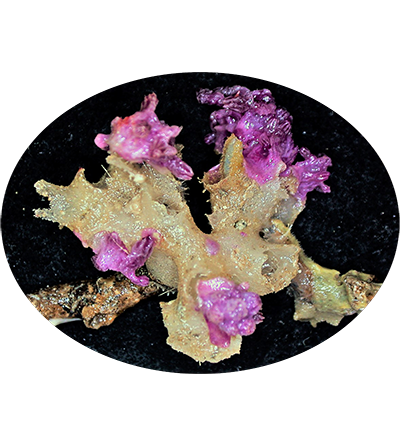
Cold-water corals Trachythela sp. YZ-2020 are important habitats for creatures in the deep-sea environment, but they have been degraded by anthropogenic activity. They live in the cold, dark, and hypoxic deep-sea waters and are widespread around the world. Most of them must attach to hard-bottom substrates to grow, and only a few can live on soft sediments. Due to the lack of symbiotic zooxanthellae, they do not require sunlight as a source of energy, and their major source of nutrients is the microscopic zooplankton that comes from passing currents or descends from the surface of the ocean.
Animalia Kingdom; Cnidaria (Phylum); Anthozoa (Class); Malacalcyonacea (Order); Clavulariidae (Family); Trachythela (Genus); Trachythela sp. YZ-2020 (Species)
Trachythela sp. YZ-2020
| Species | Phylum | Common Name | Ecosystem | Depth | Habitat | NCBI Taxonomy ID |
|---|
| Trachythela sp. YZ-2020 | Cnidaria | Trachythela sp., Cold-water corals (CWCs) | Cold seep/Hydrothermal vent | ~1068 | slope of the Xisha Trough (18°53′N, 112°66′E, July 2019) in the South China Sea (SCS) | 2770514 |
| Genome Assembly | Genome Size | Assembly level | Released year | WGS accession | Submitter | BioProject | BUSCO completeness (%) | Scaffold/Contig N50 (kb) | GC content (%) | Repeat Rate (%) | Gene Number |
|---|
| IDSSE_Tra_sp_1.0 | 578.2Mb | Contig | 2020 | JADLSH01 | Institute of Deep-Sea Science and Engineering,Chinese Academy of Sciences | PRJNA661975 | 90.70 | 3563 | 37.5 | 57.88 | 35,305 |
| Title | Journal | Pubmed ID |
|---|
| The First Draft Genome of a Cold-Water Coral Trachythela sp. (Alcyonacea: Stolonifera: Clavulariidae) | Genome Biology and Evolution | 33331875 |
| Annotation ID | Description | Type | Subtype | GeneRatio | Evolution Type | P-value | Q-value |
|---|
| GO:0004842 | ubiquitin-protein transferase activity | GO | Molecular Function | - | expanded | - | 1.14E-118 |
| GO:0006310 | DNA recombination | GO | Biological Process | - | expanded | - | 7.06E-64 |
| GO:0003676 | nucleic acid binding | GO | Molecular Function | - | expanded | - | 5.89E-55 |
| GO:0005515 | protein binding | GO | Molecular Function | - | expanded | - | 1.91E-42 |
| GO:0015074 | DNA integration | GO | Biological Process | - | expanded | - | 1.81E-40 |
| GO:0003677 | DNA binding | GO | Molecular Function | - | expanded | - | 1.76E-36 |
| GO:0016021 | integral component of membrane | GO | Cellular Component | - | expanded | - | 3.78E-34 |
| GO:0006281 | DNA repair | GO | Biological Process | - | expanded | - | 8.38E-34 |
| GO:0000723 | telomere maintenance | GO | Biological Process | - | expanded | - | 6.14E-29 |
| GO:0046983 | protein dimerization activity | GO | Molecular Function | - | expanded | - | 8.83E-27 |
| GO:0003678 | DNA helicase activity | GO | Molecular Function | - | expanded | - | 5.75E-25 |
| GO:0006313 | transposition, DNA-mediated | GO | Biological Process | - | expanded | - | 1.14E-21 |
| GO:0016020 | membrane | GO | Cellular Component | - | expanded | - | 1.90E-17 |
| GO:0016020 | membrane | GO | Cellular Component | - | contracted | - | 6.38E-08 |
| GO:0004803 | transposase activity | GO | Molecular Function | - | expanded | - | 3.81E-17 |
| GO:0055085 | transmembrane transport | GO | Biological Process | - | expanded | - | 1.30E-15 |
| GO:0003824 | catalytic activity | GO | Molecular Function | - | expanded | - | 9.79E-13 |
| GO:0007186 | G protein-coupled receptor signaling pathway | GO | Biological Process | - | expanded | - | 7.56E-11 |
| GO:0004930 | G protein-coupled receptor activity | GO | Molecular Function | - | expanded | - | 1.10E-10 |
| GO:0055114 | oxidation-reduction process | GO | Biological Process | - | expanded | - | 7.07E-10 |
| GO:0035312 | 5'-3' exodeoxyribonuclease activity | GO | Molecular Function | - | expanded | - | 7.47E-10 |