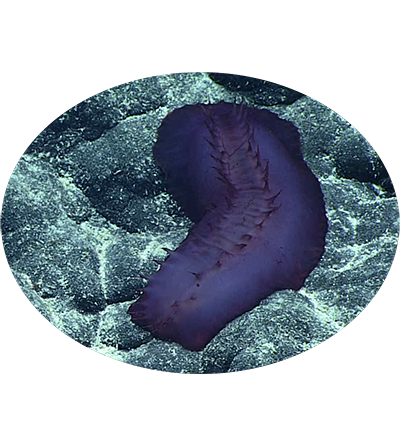
The hadal sea cucumber Paelopatides sp. Yap is a species of hadal sea cucumber in the family Stichopodidae, collected from the Yap Trench in the western Pacific Ocean. This species is adapted to the extreme conditions of the hadal zone, thriving at great depths where pressures are immense and temperatures are near freezing. Its discovery contributes to our understanding of biodiversity and ecological dynamics in one of the least explored regions of the deep sea.
| Species | Phylum | Common Name | Ecosystem | Depth | Habitat | NCBI Taxonomy ID |
|---|---|---|---|---|---|---|
| Paelopatides sp. Yap | Echinodermata | Paelopatides sp. Yap | Deep sea | 6,633 | Yap Trench at 8°1′22"N; 137°50′41"E | - |
| Genome Assembly | Genome Size | Assembly level | Released year | WGS accession | Submitter | BioProject | BUSCO completeness | Gene Number |
|---|---|---|---|---|---|---|---|---|
| - | 3.7Gb | Contig | 2020 | - | Fujian Agriculture and Forestry University | PRJCA003789 | 89.40% | 57,522 |
| Title | Journal | Pubmed ID |
|---|---|---|
| The genome of a hadal sea cucumber reveals novel adaptive strategies to deep-sea environments | iScience | 36444293 |
| Gene ID | Description |
|---|---|
| Paelopatides36260 | GLUTATHIONE S-TRANSFERASE |
| Paelopatides36261 | AMYLOID PROTEIN-BINDING PROTEIN 2 |
| Paelopatides36262 | |
| Paelopatides36263 | EXPRESSED PROTEIN |
| Paelopatides36264 | TNF RECEPTOR ASSOCIATED FACTOR |
| Paelopatides36265 | FIBRILLIN-RELATED |
| Paelopatides36266 | |
| Paelopatides36267 | FIBRILLIN-RELATED |
| Paelopatides36268 | ALPHA-TECTORIN-RELATED |
| Paelopatides36269 | |
| Paelopatides36270 | 26S PROTEASOME REGULATORY SUBUNIT |
| Paelopatides36271 | REVERSE TRANSCRIPTASE DOMAIN-CONTAINING PROTEIN |
| Paelopatides36272 | G PROTEIN-COUPLED RECEPTOR-RELATED |
| Paelopatides36273 | EIF4G DOMAIN PROTEIN |
| Paelopatides36274 | LEUCINE-RICH REPEAT-CONTAINING PROTEIN 72 |
| Paelopatides36275 | PIGGYBAC TRANSPOSABLE ELEMENT-DERIVED PROTEIN 4 |
| Paelopatides36276 | PROTEIN SIDEKICK |
| Paelopatides36277 | CYTOSOLIC CARBOXYPEPTIDASE |
| Paelopatides36278 | |
| Paelopatides36279 | DED DOMAIN-CONTAINING PROTEIN |
| Paelopatides36282 | INACTIVE TYROSINE-PROTEIN KINASE 7 |
| Paelopatides36283 | PROTEIN CBG26694 |
| Paelopatides36284 | INACTIVE TYROSINE-PROTEIN KINASE 7 |
| Paelopatides36285 | |
| Paelopatides36286 | RIBONUCLEASE |
| Paelopatides36287 | FIBRINOGEN/TENASCIN/ANGIOPOEITIN |
| Paelopatides36288 | PHAGE_INTEGRASE DOMAIN-CONTAINING PROTEIN |
| Paelopatides36289 | RETINAL PROTEIN 4 |
| Paelopatides36290 | RAB GDP/GTP EXCHANGE FACTOR |
| Paelopatides36291 | |
| Paelopatides36292 | LYR MOTIF-CONTAINING PROTEIN 9 |
| Paelopatides36293 | CELL CYCLE CONTROL PROTEIN 50 |
| Paelopatides36294 | |
| Paelopatides36295 | SARCOSINE DEHYDROGENASE-RELATED |
| Paelopatides36296 | ATP-DEPENDENT RNA HELICASE |
| Paelopatides36297 | SJOEGREN SYNDROME NUCLEAR AUTOANTIGEN 1 |
| Paelopatides36298 | |
| Paelopatides36299 | |
| Paelopatides36300 | RAB GTPASE-ACTIVATING PROTEIN 1-LIKE |
| Paelopatides36301 | |
| Paelopatides36302 | |
| Paelopatides36303 | |
| Paelopatides36304 | |
| Paelopatides36305 | |
| Paelopatides36306 | |
| Paelopatides36307 | PULMONARY SURFACTANT-ASSOCIATED PROTEIN A |
| Paelopatides36308 | PULMONARY SURFACTANT-ASSOCIATED PROTEIN A |
| Paelopatides36309 | PULMONARY SURFACTANT-ASSOCIATED PROTEIN A |
| Paelopatides36310 | |
| Paelopatides36311 |

