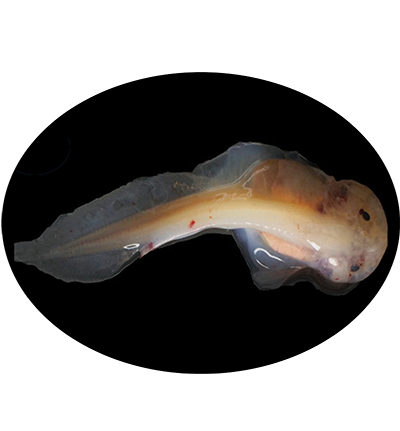
The Mariana snailfish or Mariana hadal snailfish Pseudoliparis swirei is a species of snailfish found in the Mariana Trench in the western Pacific Ocean. It is known from a depth range of 6,198–8,076 m,which is possibly the record for a fish caught on the seafloor. Various anatomical, physiological, molecular and genetic adaptions help this species survive in such depths. It is abundant in their deep-sea habitat and has several unusual adaptions for its dark and high pressure habitat, including transparent skin that lacks pigment, certain organs and eggs that are enlarged, the muscles are thinner, the ossification of its bones (notably the skull) is incomplete.
Animalia (Kingdom); Chordata (Phylum); Vertebrata (Subphylum); Gnathostomata (Infraphylum); Osteichthyes (Parvphylum); Actinopterygii (Gigaclass); Actinopteri (Superclass); Teleostei (Class); Perciformes (Order); Cottoidei (Suborder); Liparidae (Family); Pseudoliparis (Genus); Pseudoliparis swirei (Species)
Pseudoliparis swirei Gerringer & Linley, 2017
1. Gerringer M E, Linley T D, Jamieson A J, et al. Pseudoliparis swirei sp. nov.: a newly-discovered hadal snailfish (Scorpaeniformes: Liparidae) from the Mariana Trench[J]. Zootaxa, 2017, 4358(1): 161–177-161–177. (Gerringer et al., 2017)
Holotype is immature. Ripe females had eggs up to 9.4 mm diameter, among the largest teleost eggs recorded, 0.4 mm smaller than the largest record. The eggs were unsorted within gonad, with the largest eggs free and interspersed within a matrix of smaller eggs. No developmental structures were visible within even the largest eggs. Two distinct size classes of eggs present with up to 23 large eggs (>5 mm) and up to 851 small eggs of less than half the diameter of the larger size class. There were rarely intermediate stages (Figure 7). Individuals with only small eggs had maximum egg sizes ranging from 0.7 to 1.4 mm. Genital papilla visible in freshly collected males, oriented anteriorly
The Mariana Trench famously houses the ocean’s deepest point, at Challenger Deep, named for the HMS Challenger expedition which discovered the trench in 1875. Their deepest sounding of 8,184 m, then the greatest known ocean depth, was christened Swire Deep after Herbert Swire, the ship’s First Navigating Sublieutenant (Corfield 2003). We name this fish in his honor, in acknowledgment and gratitude of the crew members that have supported oceanographic research throughout history
Known only from the Mariana Trench at capture depths from 6,898–7,966 m, individuals likely this species were recognized in video at depths 6,198–8,098 m
| Species | Phylum | Common Name | Ecosystem | Depth | Habitat | NCBI Taxonomy ID |
|---|---|---|---|---|---|---|
| Pseudoliparis swirei | Chordata | hadal snailfish | Deep sea | 7,254 | the Mariana Trench (142°26′E, 11°07′N) | 2059687 |
| Genome Assembly | Genome Size | Assembly level | Released year | WGS accession | Submitter | BioProject | BUSCO completeness (%) | Scaffold/Contig N50 (kb) | GC content (%) | Repeat Rate (%) | Gene Number |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NWPU_hadal_v1 | 626.4Mb | Chromosome | 2023 | JANBZZ01 | Northwestern Polytechnical University | PRJNA852951 | 96.00 | 25,738/4,219 | 44 | - | 21,161 |
| Title | Journal | Pubmed ID |
|---|---|---|
| Chromosome-level genome assembly of hadal snailfish reveals mechanisms of deep-sea adaptation in vertebrates | Elife | 38134226 |
| Gene ID | Description |
|---|---|
| HS02281 | ATP-DEPENDENT RNA HELICASE |
| HS02282 | P2X PURINOCEPTOR |
| HS02283 | MAP/MICROTUBULE AFFINITY-REGULATING KINASE |
| HS02284 | CALCIUM-TRANSPORTING ATPASE |
| HS02285 | NOVEL ZZ TYPE ZINC FINGER DOMAIN CONTAINING PROTEIN |
| HS02286 | MEMBRANE-ASSOCIATED PROGESTERONE RECEPTOR COMPONENT-RELATED |
| HS02287 | POTE ANKYRIN DOMAIN |
| HS02288 | UBIQUITIN-CONJUGATING ENZYME E2 |
| HS02289 | UNCHARACTERIZED RING FINGER-CONTAINING PROTEIN |
| HS02290 | RIBOSOMAL PROTEIN S6 KINASE |
| HS02291 | MEMBRANE-ASSOCIATED PROGESTERONE RECEPTOR COMPONENT-RELATED |
| HS02292 | PEPTIDYL-TRNA HYDROLASE 2 |
| HS02293 | CLATHRIN HEAVY CHAIN RELATED |
| HS02294 | PDZ DOMAIN CONTAINING PROTEIN |
| HS02295 | ATP SYNTHASE SUBUNIT |
| HS02296 | B-BOX DOMAIN CONTAINING |
| HS02297 | B-BOX DOMAIN CONTAINING |
| HS02298 | RUVB-RELATED REPTIN AND PONTIN |
| HS02299 | - |
| HS02300 | SPINSTER |

