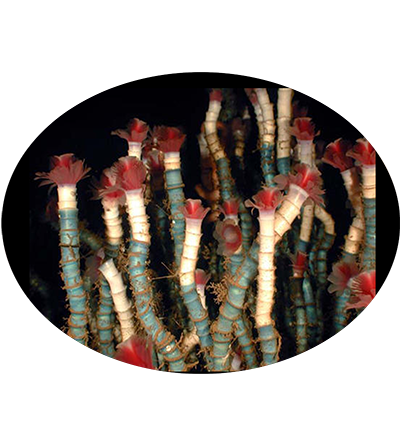
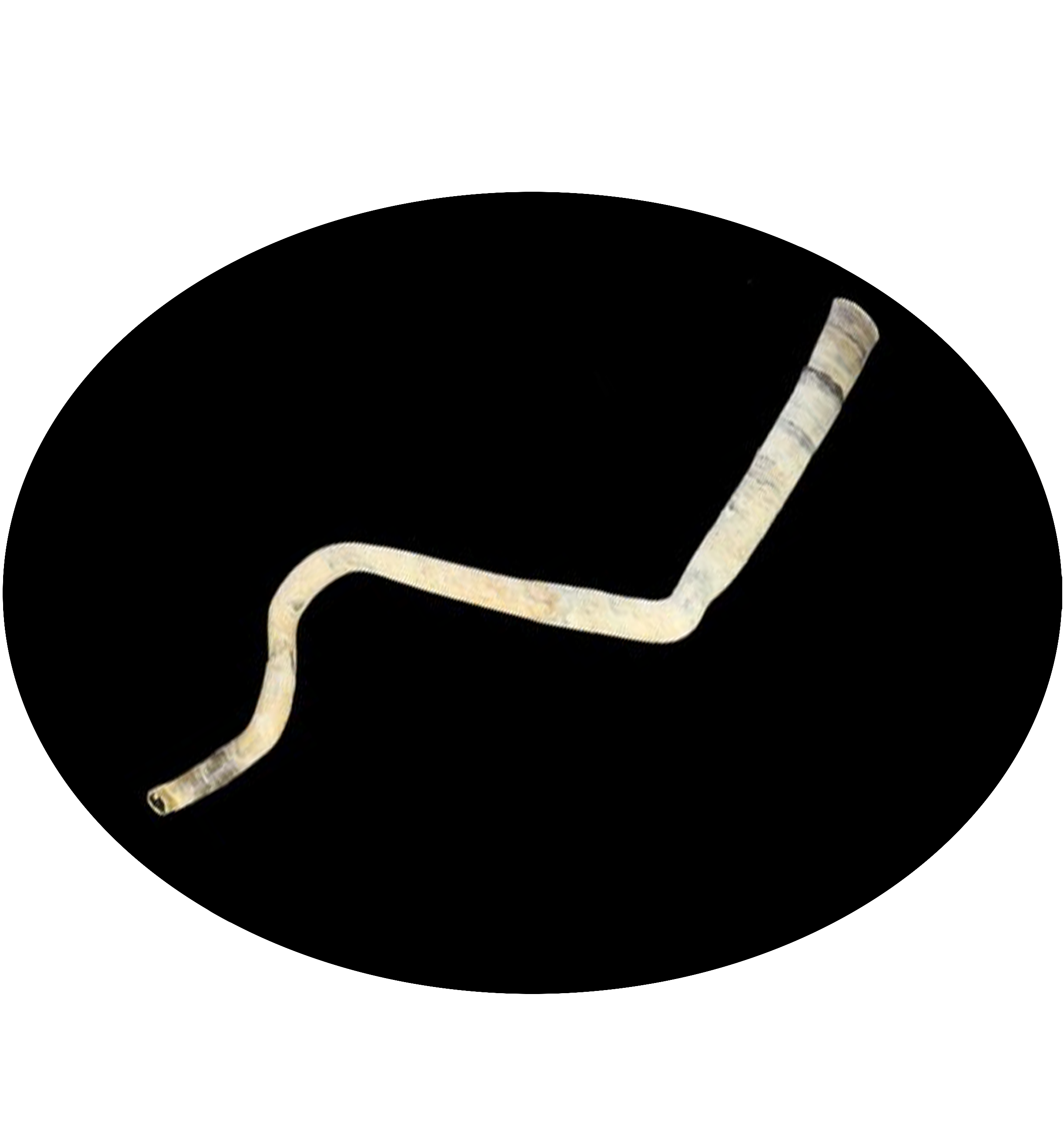
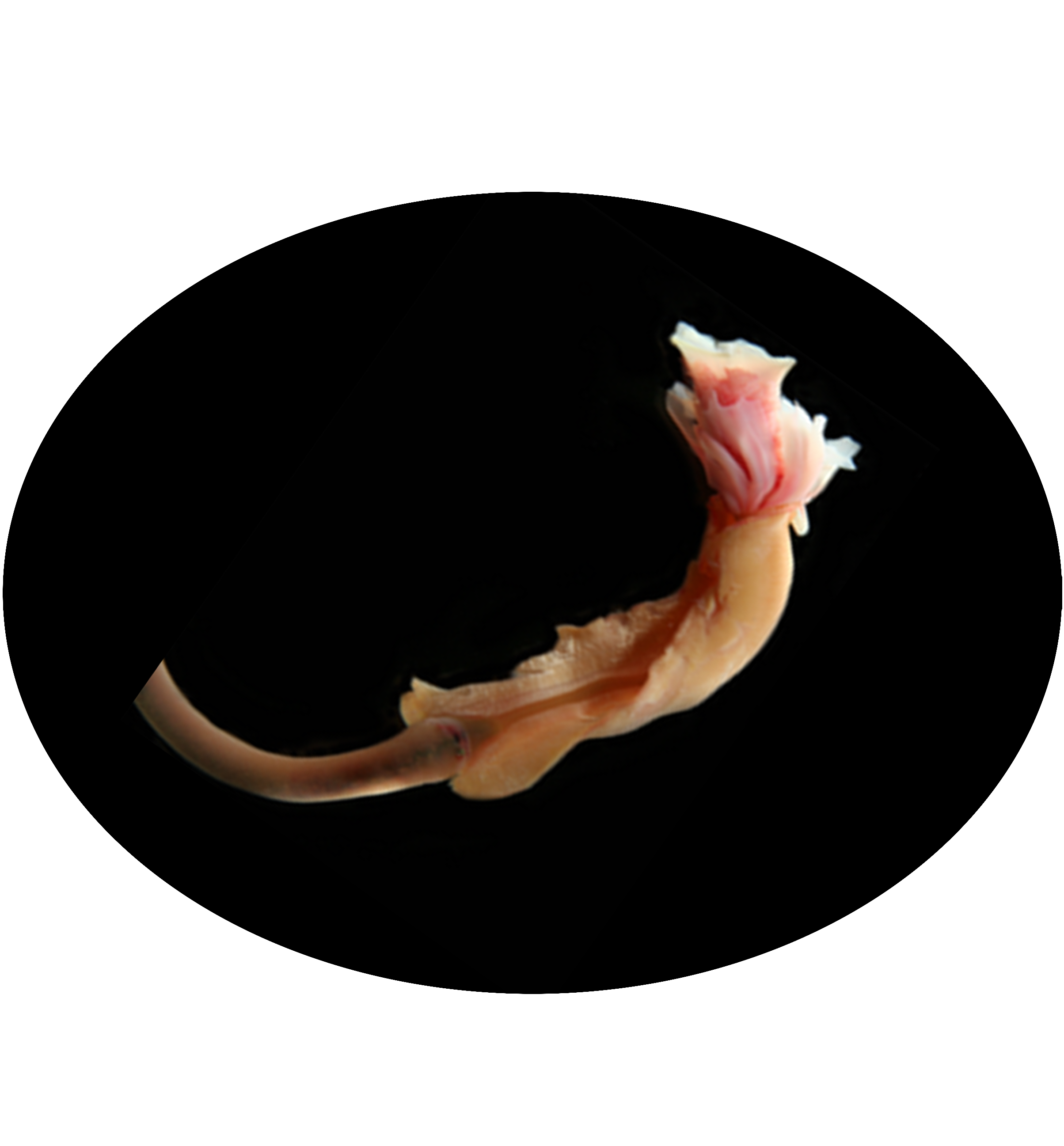
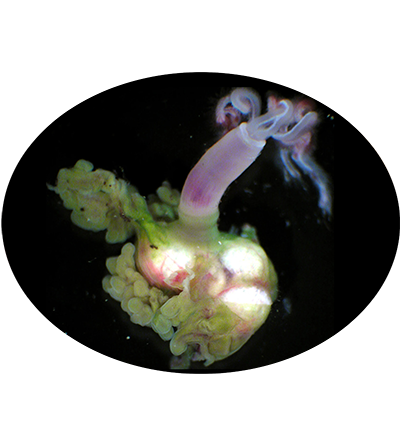
Welcome to DOO!
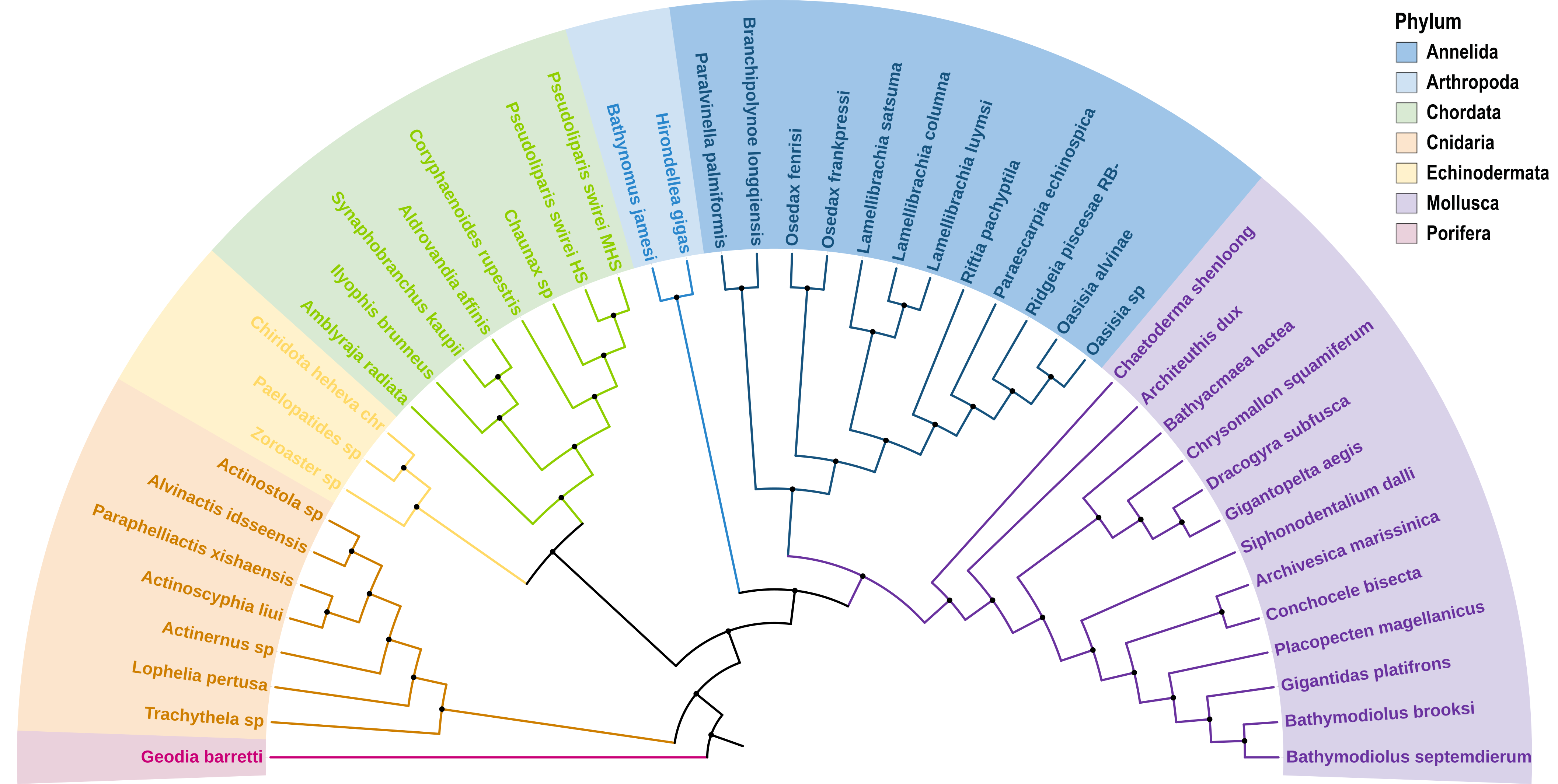
The deep ocean is one of Earth’s most vast and least explored frontiers, characterised by extreme conditions such as high pressure, limited light and nutrient scarcity. These environments pose unparalleled challenges to life, making them invaluable for studying genetic and molecular adaptations to extreme conditions. Emerging omics resources have recently provided significant insights into the advanced understanding of deep ocean ecosystems and evolution. However, a centralised resource for deep ocean multi-omics data remains lacking. To bridge this gap, we present the Deep Ocean Omics (DOO) database, a multi-omics atlas for deep ocean organisms. DOO integrates diverse omics resources from 68 species across seven phyla and 16 classes, encompassing 72 genomes, 950 bulk transcriptomes, 15 single-cell transcriptomes and 1112 metagenomes, alongside functional support toolkits for functional and comparative analysis. DOO provides a systematic view of genomic information, including genome assembly, phylogeny, gene annotation, BUSCO genes, transcription factors/ubiquitin family, gene cluster, symbiont and mitochondrial genomes and fossil records. Moreover, DOO offers co-expression networks with expression views across different tissues and developmental stages and micro- and macrosynteny analyses to elucidate the pan-evolutionary features of genome structure. As the first comprehensive multi-omics resource dedicated to deep ocean organisms, DOO serves as a pivotal platform for uncovering multi-omics underpinnings of deep ocean organisms and offering insights into the understanding of deep ocean biodiversity, evolution and genetic adaptation under extreme conditions.
Choose Species >>>
Contact Us
If you have any questions, comments or suggestions about our website or our project, please feel free to contact us: jackieyihkb@ust.hk, longjunwu@ust.hk and boqianpy@ust.hk. If you want to know more about our work, welcome to Longjun Wu's Lab and Pei-Yuan Qian's Lab.
Cite Us
Jiajie She, Pei-yuan Qian, Longjun Wu, DOO: integrated multi-omics resources for deep ocean organisms, Nucleic Acids Res 2025. https://doi.org/10.1093/nar/gkaf1096.
- Phylum/Class/Order/Speices: 7/15/35/68
- Fossil records: 1,413
- Protein-coding genes: 1,449,954
- Busco Genes: 16,015
- Transcriptomic datasets: 950
- Co-expressed gene pairs:2,005,358
- SnRNA-seq datasets:15
- Cell types:14
- Cell markers:1,794
- Metagenome datasets:1,112
- Assembled symbiont genomes: 131
- Annotated microbial genes: 92,682
- Mitochondrial datasets: 53
- Type of gene annotation:7
- KEGG pathways: 18,539
- Transcription factors: 60,866
- Ubiquitin family: 82,844
- Gene Ontology: 3,606,859
- Interpro Annotation: 3,640,096
- Pfam Domain: 1,281,713
- PATHER: 928,123
- Macrosynteny gene pairs: 398,247
- Gene family: 1,385,060
- The Ocean Decade
- Mariana Trench Environment and Ecology Research (MEER)
- Deep Ocean Microbiomes and Ecosystems (DOME)
- World Register of Marine species (WoRMS)
- World Register of Deep-Sea Species (WoRDDS)
- Global Biodiversity Information Facility (GBIF)
- The Paleobiology Database (PBDB)
- SeaLifeBase
- Earth BioGenome Project
- Planet Microbe
- 2025-07-09: DOO is online now!
- 2025-05-14: Single-cell transcriptomic analysis module including cell atlas, cell marker and gene expression is also available!
- 2025-04-19: Microsynteny Analysis is provided!
- 2025-04-02: The information of Busco genes is provided!
- 2025-02-23: Macrosynteny Analysis is provided!
- 2025-01-22: Single-cell transcriptomic data module was developed.
- 2024-12-13: Mitogenomic Data is available!
- 2024-11-26: Symbiotic Microorganism Data is provided!
- 2024-08-08: Dynamic expression view is available!
- 2024-05-30: The part of Data Submit & Comments is available!
- 2024-05-22: Network Search and Analysis is provided!
- 2024-05-20: Gene Sets Enrichment Analysis is available!
- 2024-05-03: Blast Alignment is available!
- 2024-04-18: Initial platform framework of DOO is online!