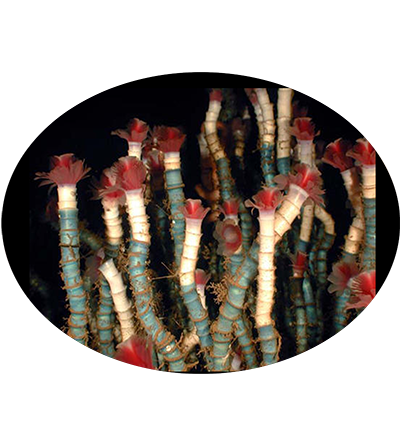
Lamellibrachia luymesi is a species of tube worms in the family Siboglinidae. It lives at deep-sea cold seeps where hydrocarbons (oil and methane) are leaking out of the seafloor. It is entirely reliant on internal, sulfide-oxidizing bacterial symbionts for its nutrition. These are located in a centrally located 'trophosome'. It provides the bacteria with hydrogen sulfide and oxygen by taking them up from the environment and binding them to a specialized hemoglobin molecule. It may help fuel the generation of sulfide by excreting sulfate through their roots into the sediments below the aggregations.
| Species | Phylum | Common Name | Ecosystem | Depth | Habitat | NCBI Taxonomy ID |
|---|---|---|---|---|---|---|
| Lamellibrachia luymesi | Annelida | cold seep-dwelling tubeworm Lamellibrachia luymesi | Hydrothermal vent | >540 | the Gulf of Mexico (N 28° 11.58′, W 89° 47.94′) | 238240 |
| Genome Assembly | Genome Size | Assembly level | Released year | WGS accession | Submitter | BioProject | BUSCO completeness | Gene Number |
|---|---|---|---|---|---|---|---|---|
| LLUY_1.0 | ~688Mb | Scaffold | 2019 | SDWI01 | Auburn University | PRJNA516467 | ~95% | 38,998 |
| Title | Journal | Pubmed ID |
|---|---|---|
| Genomic adaptations to chemosymbiosis in the deep-sea seep-dwelling tubeworm Lamellibrachia luymesi | BMC Biology | 31739792 |
| Gene ID | Description |
|---|---|
| LLU|FUN_019311-T1 | |
| LLU|FUN_019312-T1 | METHYLMALONYL-COA EPIMERASE |
| LLU|FUN_019313-T1 | ANGIOGENIC FACTOR WITH G PATCH AND FHA DOMAINS 1 |
| LLU|FUN_019314-T1 | |
| LLU|FUN_019315-T1 | |
| LLU|FUN_019316-T1 | |
| LLU|FUN_019317-T1 | |
| LLU|FUN_019318-T1 | TOLL-LIKE RECEPTOR |
| LLU|FUN_019319-T1 | NUCLEOLAR RNA-ASSOCIATED PROTEIN |
| LLU|FUN_019320-T1 | |
| LLU|FUN_019321-T1 | |
| LLU|FUN_019322-T1 | |
| LLU|FUN_019323-T1 | PHOSPHORYLASE B KINASE REGULATORY SUBUNIT |
| LLU|FUN_019324-T1 | GRB2-ASSOCIATED AND REGULATOR OF MAPK PROTEIN FAMILY MEMBER |
| LLU|FUN_019325-T1 | L-FUCOSE KINASE |
| LLU|FUN_019326-T1 | GRB2-ASSOCIATED AND REGULATOR OF MAPK PROTEIN FAMILY MEMBER |
| LLU|FUN_019327-T1 | |
| LLU|FUN_019328-T1 | T-CELL IMMUNOMODULATORY PROTEIN HOMOLOG |
| LLU|FUN_019329-T1 | |
| LLU|FUN_019330-T1 | T-CELL IMMUNOMODULATORY PROTEIN HOMOLOG |
| LLU|FUN_019331-T1 | G PATCH DOMAIN-CONTAINING PROTEIN 1 |
| LLU|FUN_019332-T1 | PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY SUBUNIT |
| LLU|FUN_019333-T1 | UBIQUINONE BIOSYNTHESIS PROTEIN COQ9, MITOCHONDRIAL |
| LLU|FUN_019334-T1 | GLOMULIN |
| LLU|FUN_019335-T1 | LEUCINE-RICH REPEAT, ISOFORM F-RELATED |
| LLU|FUN_019336-T1 | |
| LLU|FUN_019338-T1 | |
| LLU|FUN_019339-T1 | TRANSPORT PROTEIN TRAPP |
| LLU|FUN_019340-T1 | |
| LLU|FUN_019341-T1 | ANKYRIN REPEAT, SAM AND BASIC LEUCINE ZIPPER DOMAIN-CONTAINING PROTEIN 1 |
| LLU|FUN_019342-T1 | |
| LLU|FUN_019343-T1 | |
| LLU|FUN_019344-T1 | IRE1-RELATED |
| LLU|FUN_019345-T1 | |
| LLU|FUN_019346-T1 | SOLUTE CARRIER FAMILY 52 |
| LLU|FUN_019347-T1 | |
| LLU|FUN_019348-T1 | |
| LLU|FUN_019349-T1 | |
| LLU|FUN_019350-T1 | |
| LLU|FUN_019351-T1 | |
| LLU|FUN_019352-T1 | METHYLTHIOADENOSINE/PURINE NUCLEOSIDE PHOSPHORYLASE |
| LLU|FUN_019353-T1 | METHYLTHIOADENOSINE/PURINE NUCLEOSIDE PHOSPHORYLASE |
| LLU|FUN_019354-T1 | BLR2807 PROTEIN |
| LLU|FUN_019355-T1 | ADENYLATE CYCLASE TYPE 1 |
| LLU|FUN_019356-T1 | ADENYLATE CYCLASE TYPE 1 |
| LLU|FUN_019357-T1 | METHYLTHIOADENOSINE/PURINE NUCLEOSIDE PHOSPHORYLASE |
| LLU|FUN_019358-T1 | ANKYRIN REPEAT DOMAIN-CONTAINING PROTEIN 60 |
| LLU|FUN_019359-T1 | |
| LLU|FUN_019360-T1 | EUKARYOTIC TRANSLATION INITIATION FACTOR 3 -RELATED |
| LLU|FUN_019361-T1 | UNCHARACTERIZED |

