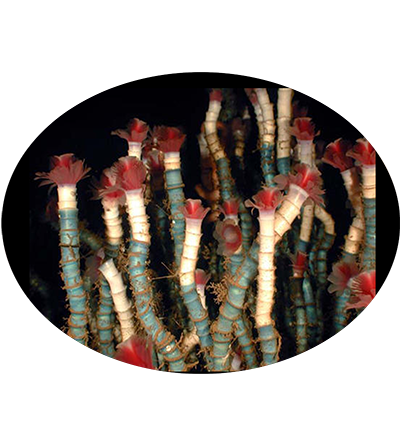
Lamellibrachia luymesi is a species of tube worms in the family Siboglinidae. It lives at deep-sea cold seeps where hydrocarbons (oil and methane) are leaking out of the seafloor. It is entirely reliant on internal, sulfide-oxidizing bacterial symbionts for its nutrition. These are located in a centrally located 'trophosome'. It provides the bacteria with hydrogen sulfide and oxygen by taking them up from the environment and binding them to a specialized hemoglobin molecule. It may help fuel the generation of sulfide by excreting sulfate through their roots into the sediments below the aggregations.
| Species | Phylum | Common Name | Ecosystem | Depth | Habitat | NCBI Taxonomy ID |
|---|---|---|---|---|---|---|
| Lamellibrachia luymesi | Annelida | cold seep-dwelling tubeworm Lamellibrachia luymesi | Hydrothermal vent | >540 | the Gulf of Mexico (N 28° 11.58′, W 89° 47.94′) | 238240 |
| Genome Assembly | Genome Size | Assembly level | Released year | WGS accession | Submitter | BioProject | BUSCO completeness | Gene Number |
|---|---|---|---|---|---|---|---|---|
| LLUY_1.0 | ~688Mb | Scaffold | 2019 | SDWI01 | Auburn University | PRJNA516467 | ~95% | 38,998 |
| Title | Journal | Pubmed ID |
|---|---|---|
| Genomic adaptations to chemosymbiosis in the deep-sea seep-dwelling tubeworm Lamellibrachia luymesi | BMC Biology | 31739792 |
| Gene ID | Description |
|---|---|
| LLU|FUN_030611-T1 | NEUROTRANSMITTER GATED ION CHANNEL |
| LLU|FUN_030612-T1 | |
| LLU|FUN_030613-T1 | |
| LLU|FUN_030614-T1 | ADENOSINE DEAMINASE |
| LLU|FUN_030615-T1 | MUCOSA ASSOCIATED LYMPHOID TISSUE LYMPHOMA TRANSLOCATION PROTEIN 1/PARACASPASE |
| LLU|FUN_030616-T1 | HAT FAMILY DIMERISATION DOMAINCONTAINING PROTEIN-RELATED |
| LLU|FUN_030617-T1 | MACROGLOBULIN / COMPLEMENT |
| LLU|FUN_030618-T1 | MACROGLOBULIN / COMPLEMENT |
| LLU|FUN_030619-T1 | |
| LLU|FUN_030620-T1 | |
| LLU|FUN_030621-T1 | |
| LLU|FUN_030622-T1 | |
| LLU|FUN_030623-T1 | FI19480P1 |
| LLU|FUN_030624-T1 | PEPTIDYL-TRNA HYDROLASE PTRHD1-RELATED |
| LLU|FUN_030625-T1 | |
| LLU|FUN_030626-T1 | |
| LLU|FUN_030627-T1 | |
| LLU|FUN_030628-T1 | 3-KETO-STEROID REDUCTASE |
| LLU|FUN_030629-T1 | REGULATOR OF MICROTUBULE DYNAMICS PROTEIN |
| LLU|FUN_030630-T1 | |
| LLU|FUN_030631-T1 | SPHINGOSINE KINASE |
| LLU|FUN_030632-T1 | MSL2-RELATED |
| LLU|FUN_030633-T1 | |
| LLU|FUN_030634-T1 | ANKYRIN REPEAT DOMAIN-CONTAINING PROTEIN 39-RELATED |
| LLU|FUN_030635-T1 | |
| LLU|FUN_030636-T1 | |
| LLU|FUN_030637-T1 | HEPARAN SULFATE 2-O-SULFOTRANSFERASE |
| LLU|FUN_030638-T1 | PROTEIN EVA-1 |
| LLU|FUN_030639-T1 | |
| LLU|FUN_030641-T1 | |
| LLU|FUN_030642-T1 | |
| LLU|FUN_030643-T1 | |
| LLU|FUN_030644-T1 | |
| LLU|FUN_030645-T1 | |
| LLU|FUN_030646-T1 | |
| LLU|FUN_030647-T1 | INDOLEAMINE 2,3-DIOXYGENASE |
| LLU|FUN_030648-T1 | |
| LLU|FUN_030649-T1 | INDOLEAMINE 2,3-DIOXYGENASE |
| LLU|FUN_030650-T1 | PROTEASE FAMILY C26 GAMMA-GLUTAMYL HYDROLASE |
| LLU|FUN_030651-T1 | CXC DOMAIN-CONTAINING PROTEIN-RELATED |
| LLU|FUN_030652-T1 | CXC DOMAIN-CONTAINING PROTEIN-RELATED |
| LLU|FUN_030653-T1 | HAT FAMILY DIMERISATION DOMAINCONTAINING PROTEIN-RELATED |
| LLU|FUN_030654-T1 | |
| LLU|FUN_030655-T1 | PROTEASE FAMILY C26 GAMMA-GLUTAMYL HYDROLASE |
| LLU|FUN_030656-T1 | |
| LLU|FUN_030657-T1 | PROTEASE FAMILY C26 GAMMA-GLUTAMYL HYDROLASE |
| LLU|FUN_030658-T1 | CALCIUM BINDING PROTEIN |
| LLU|FUN_030659-T1 | BRIDGING INTEGRATOR 3 |
| LLU|FUN_030660-T1 | UNCHARACTERIZED |
| LLU|FUN_030661-T1 | PDZ DOMAIN CONTAINING WHIRLIN AND HARMONIN-RELATED |

