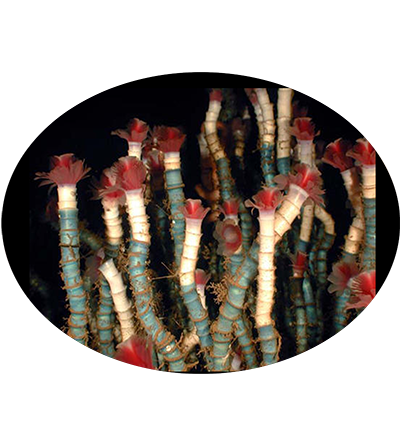
Lamellibrachia luymesi is a species of tube worms in the family Siboglinidae. It lives at deep-sea cold seeps where hydrocarbons (oil and methane) are leaking out of the seafloor. It is entirely reliant on internal, sulfide-oxidizing bacterial symbionts for its nutrition. These are located in a centrally located 'trophosome'. It provides the bacteria with hydrogen sulfide and oxygen by taking them up from the environment and binding them to a specialized hemoglobin molecule. It may help fuel the generation of sulfide by excreting sulfate through their roots into the sediments below the aggregations.
| Species | Phylum | Common Name | Ecosystem | Depth | Habitat | NCBI Taxonomy ID |
|---|---|---|---|---|---|---|
| Lamellibrachia luymesi | Annelida | cold seep-dwelling tubeworm Lamellibrachia luymesi | Hydrothermal vent | >540 | the Gulf of Mexico (N 28° 11.58′, W 89° 47.94′) | 238240 |
| Genome Assembly | Genome Size | Assembly level | Released year | WGS accession | Submitter | BioProject | BUSCO completeness | Gene Number |
|---|---|---|---|---|---|---|---|---|
| LLUY_1.0 | ~688Mb | Scaffold | 2019 | SDWI01 | Auburn University | PRJNA516467 | ~95% | 38,998 |
| Title | Journal | Pubmed ID |
|---|---|---|
| Genomic adaptations to chemosymbiosis in the deep-sea seep-dwelling tubeworm Lamellibrachia luymesi | BMC Biology | 31739792 |
| Gene ID | Description |
|---|---|
| LLU|FUN_030409-T1 | ARMADILLO REPEAT-CONTAINING PROTEIN 1 |
| LLU|FUN_030410-T1 | ADP-RIBOSYLATION FACTOR GTPASE-ACTIVATING PROTEIN 1 |
| LLU|FUN_030411-T1 | DUAL SPECIFICITY PROTEIN PHOSPHATASE |
| LLU|FUN_030412-T1 | RNA 3' TERMINAL PHOSPHATE CYCLASE |
| LLU|FUN_030413-T1 | DNA EXCISION REPAIR PROTEIN ERCC-8 |
| LLU|FUN_030414-T1 | HISTONE DEACETYLASE HDAC1-RELATED |
| LLU|FUN_030415-T1 | GLYCINE DEHYDROGENASE, DECARBOXYLATING |
| LLU|FUN_030416-T1 | NARDILYSIN |
| LLU|FUN_030417-T1 | SOLUTE CARRIER FAMILY 17 |
| LLU|FUN_030418-T1 | |
| LLU|FUN_030419-T1 | |
| LLU|FUN_030420-T1 | |
| LLU|FUN_030421-T1 | PROTEIN DISULFIDE-ISOMERASE C17H9.14C-RELATED |
| LLU|FUN_030422-T1 | |
| LLU|FUN_030423-T1 | |
| LLU|FUN_030424-T1 | KRUEPPEL-LIKE TRANSCRIPTION FACTOR |
| LLU|FUN_030425-T1 | COBW-RELATED |
| LLU|FUN_030426-T1 | |
| LLU|FUN_030427-T1 | |
| LLU|FUN_030428-T1 | |
| LLU|FUN_030429-T1 | |
| LLU|FUN_030430-T1 | FORKHEAD BOX PROTEIN |
| LLU|FUN_030431-T1 | |
| LLU|FUN_030432-T1 | |
| LLU|FUN_030433-T1 | |
| LLU|FUN_030434-T1 | |
| LLU|FUN_030435-T1 | MOLTING PROTEIN MLT-4 |
| LLU|FUN_030436-T1 | ALPHA-GLUCOSIDASE |
| LLU|FUN_030437-T1 | ALPHA-GLUCOSIDASE |
| LLU|FUN_030438-T1 | |
| LLU|FUN_030439-T1 | INACTIVE POLYGLYCYLASE TTLL10 |
| LLU|FUN_030440-T1 | PROTEIN MAGO NASHI HOMOLOG |
| LLU|FUN_030441-T1 | SYNAPTIC VESICLE GLYCOPROTEIN 2 |
| LLU|FUN_030442-T1 | |
| LLU|FUN_030443-T1 | - |
| LLU|FUN_030444-T1 | PCNA-INTERACTING PARTNER |
| LLU|FUN_030445-T1 | MRG-BINDING PROTEIN |
| LLU|FUN_030446-T1 | FMRFAMIDE RECEPTOR-RELATED |
| LLU|FUN_030447-T1 | |
| LLU|FUN_030448-T1 | |
| LLU|FUN_030449-T1 | ALL-TRANS RETINOIC ACID-INDUCED DIFFERENTIATION FACTOR |
| LLU|FUN_030450-T1 | TEKTIN |
| LLU|FUN_030451-T1 | |
| LLU|FUN_030452-T1 | FORKHEAD BOX PROTEIN |
| LLU|FUN_030453-T1 | |
| LLU|FUN_030454-T1 | FORKHEAD BOX PROTEIN |
| LLU|FUN_030455-T1 | ARGININE/DIAMINOPIMELATE/ORNITHINE DECARBOXYLASE |
| LLU|FUN_030456-T1 | ARGININE/DIAMINOPIMELATE/ORNITHINE DECARBOXYLASE |
| LLU|FUN_030457-T1 | RNA POLYMERASE-ASSOCIATED PROTEIN CTR9 |
| LLU|FUN_030458-T1 | SIGNAL PEPTIDE, CUB AND EGF-LIKE DOMAIN-CONTAINING |

