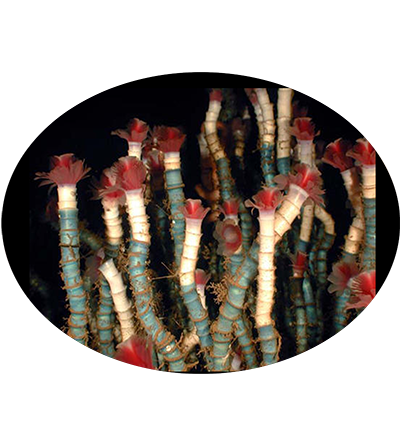
Lamellibrachia luymesi is a species of tube worms in the family Siboglinidae. It lives at deep-sea cold seeps where hydrocarbons (oil and methane) are leaking out of the seafloor. It is entirely reliant on internal, sulfide-oxidizing bacterial symbionts for its nutrition. These are located in a centrally located 'trophosome'. It provides the bacteria with hydrogen sulfide and oxygen by taking them up from the environment and binding them to a specialized hemoglobin molecule. It may help fuel the generation of sulfide by excreting sulfate through their roots into the sediments below the aggregations.
| Species | Phylum | Common Name | Ecosystem | Depth | Habitat | NCBI Taxonomy ID |
|---|---|---|---|---|---|---|
| Lamellibrachia luymesi | Annelida | cold seep-dwelling tubeworm Lamellibrachia luymesi | Hydrothermal vent | >540 | the Gulf of Mexico (N 28° 11.58′, W 89° 47.94′) | 238240 |
| Genome Assembly | Genome Size | Assembly level | Released year | WGS accession | Submitter | BioProject | BUSCO completeness | Gene Number |
|---|---|---|---|---|---|---|---|---|
| LLUY_1.0 | ~688Mb | Scaffold | 2019 | SDWI01 | Auburn University | PRJNA516467 | ~95% | 38,998 |
| Title | Journal | Pubmed ID |
|---|---|---|
| Genomic adaptations to chemosymbiosis in the deep-sea seep-dwelling tubeworm Lamellibrachia luymesi | BMC Biology | 31739792 |
| Gene ID | Description |
|---|---|
| LLU|FUN_016071-T1 | |
| LLU|FUN_016072-T1 | RGD1565685 PROTEIN |
| LLU|FUN_016073-T1 | UNCHARACTERIZED |
| LLU|FUN_016074-T1 | GLUCOSE-1-PHOSPHATE THYMIDYLTRANSFERASE |
| LLU|FUN_016075-T1 | RYANODINE RECEPTOR AND IP3 RECEPTOR |
| LLU|FUN_016076-T1 | LIPOPOLYSACCHARIDE CHOLINEPHOSPHOTRANSFERASE LICD |
| LLU|FUN_016077-T1 | |
| LLU|FUN_016078-T1 | BASIC HELIX-LOOP-HELIX PROTEIN NEUROGENIN-RELATED |
| LLU|FUN_016079-T1 | |
| LLU|FUN_016080-T1 | |
| LLU|FUN_016081-T1 | |
| LLU|FUN_016082-T1 | RGD1565685 PROTEIN |
| LLU|FUN_016083-T1 | TOLL-LIKE RECEPTOR |
| LLU|FUN_016084-T1 | |
| LLU|FUN_016085-T1 | |
| LLU|FUN_016086-T1 | |
| LLU|FUN_016087-T1 | |
| LLU|FUN_016088-T1 | PROTEIN CBG12054-RELATED |
| LLU|FUN_016089-T1 | |
| LLU|FUN_016090-T1 | |
| LLU|FUN_016091-T1 | |
| LLU|FUN_016092-T1 | |
| LLU|FUN_016093-T1 | |
| LLU|FUN_016094-T1 | |
| LLU|FUN_016095-T1 | ELYS PROTEIN |
| LLU|FUN_016096-T1 | NUCLEAR RECEPTOR SUBFAMILY 0 GROUP B |
| LLU|FUN_016097-T1 | |
| LLU|FUN_016098-T1 | |
| LLU|FUN_016099-T1 | - |
| LLU|FUN_016100-T1 | TRANSMEMBRANE PROTEIN 208 |
| LLU|FUN_016101-T1 | CHST5 PROTEIN |
| LLU|FUN_016102-T1 | |
| LLU|FUN_016103-T1 | PROTEIN-GLUTAMINE GAMMA-GLUTAMYLTRANSFERASE |
| LLU|FUN_016104-T1 | |
| LLU|FUN_016105-T1 | PROTEIN-GLUTAMINE GAMMA-GLUTAMYLTRANSFERASE |
| LLU|FUN_016106-T1 | MITOCHONDRIAL IMPORT INNER MEMBRANE TRANSLOCASE SUBUNIT TIM21 |
| LLU|FUN_016107-T1 | U3 SMALL NUCLEOLAR RNA-ASSOCIATED PROTEIN 4 HOMOLOG |
| LLU|FUN_016108-T1 | IMPORTIN BETA |
| LLU|FUN_016109-T1 | ENDONUCLEASE/EXONUCLEASE/PHOSPHATASE FAMILY DOMAIN-CONTAINING PROTEIN 1 |
| LLU|FUN_016110-T1 | ZINC FINGER PROTEIN |
| LLU|FUN_016111-T1 | BAX INHIBITOR-RELATED |
| LLU|FUN_016112-T1 | ATP-DEPENDENT RNA HELICASE RHLE-RELATED |
| LLU|FUN_016113-T1 | FMRFAMIDE RECEPTOR-RELATED |
| LLU|FUN_016114-T1 | ATP-DEPENDENT RNA HELICASE DBP3 |
| LLU|FUN_016115-T1 | |
| LLU|FUN_016116-T1 | |
| LLU|FUN_016117-T1 | |
| LLU|FUN_016118-T1 | POTASSIUM CHANNEL TETRAMERIZATION DOMAIN-CONTAINING |
| LLU|FUN_016119-T1 | MACROGLOBULIN / COMPLEMENT |
| LLU|FUN_016120-T1 | VOLTAGE-DEPENDENT CALCIUM CHANNEL SUBUNIT ALPHA-2/DELTA-RELATED |

