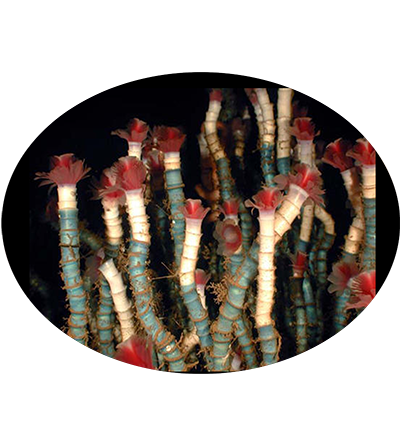
Lamellibrachia luymesi is a species of tube worms in the family Siboglinidae. It lives at deep-sea cold seeps where hydrocarbons (oil and methane) are leaking out of the seafloor. It is entirely reliant on internal, sulfide-oxidizing bacterial symbionts for its nutrition. These are located in a centrally located 'trophosome'. It provides the bacteria with hydrogen sulfide and oxygen by taking them up from the environment and binding them to a specialized hemoglobin molecule. It may help fuel the generation of sulfide by excreting sulfate through their roots into the sediments below the aggregations.
| Species | Phylum | Common Name | Ecosystem | Depth | Habitat | NCBI Taxonomy ID |
|---|---|---|---|---|---|---|
| Lamellibrachia luymesi | Annelida | cold seep-dwelling tubeworm Lamellibrachia luymesi | Hydrothermal vent | >540 | the Gulf of Mexico (N 28° 11.58′, W 89° 47.94′) | 238240 |
| Genome Assembly | Genome Size | Assembly level | Released year | WGS accession | Submitter | BioProject | BUSCO completeness | Gene Number |
|---|---|---|---|---|---|---|---|---|
| LLUY_1.0 | ~688Mb | Scaffold | 2019 | SDWI01 | Auburn University | PRJNA516467 | ~95% | 38,998 |
| Title | Journal | Pubmed ID |
|---|---|---|
| Genomic adaptations to chemosymbiosis in the deep-sea seep-dwelling tubeworm Lamellibrachia luymesi | BMC Biology | 31739792 |
| Gene ID | Description |
|---|---|
| LLU|FUN_008782-T1 | NETRIN RECEPTOR UNC5 |
| LLU|FUN_008783-T1 | NETRIN RECEPTOR UNC5 |
| LLU|FUN_008784-T1 | |
| LLU|FUN_008785-T1 | |
| LLU|FUN_008786-T1 | |
| LLU|FUN_008787-T1 | HOMEOBOX PROTEIN ENGRAILED |
| LLU|FUN_008788-T1 | |
| LLU|FUN_008790-T1 | E3 UBIQUITIN-PROTEIN LIGASE SNT2 |
| LLU|FUN_008791-T1 | EG:EG0003.4 PROTEIN-RELATED |
| LLU|FUN_008792-T1 | |
| LLU|FUN_008793-T1 | |
| LLU|FUN_008794-T1 | NEPHRIN |
| LLU|FUN_008795-T1 | HISTAMINE RECEPTOR-RELATED G-PROTEIN COUPLED RECEPTOR |
| LLU|FUN_008796-T1 | HISTAMINE RECEPTOR-RELATED G-PROTEIN COUPLED RECEPTOR |
| LLU|FUN_008797-T1 | |
| LLU|FUN_008798-T1 | |
| LLU|FUN_008799-T1 | CARTILAGE INTERMEDIATE LAYER PROTEIN CLIP |
| LLU|FUN_008800-T1 | |
| LLU|FUN_008801-T1 | CARTILAGE INTERMEDIATE LAYER PROTEIN CLIP |
| LLU|FUN_008802-T1 | CARTILAGE INTERMEDIATE LAYER PROTEIN CLIP |
| LLU|FUN_008803-T1 | CARTILAGE INTERMEDIATE LAYER PROTEIN CLIP |
| LLU|FUN_008804-T1 | |
| LLU|FUN_008805-T1 | |
| LLU|FUN_008806-T1 | |
| LLU|FUN_008807-T1 | |
| LLU|FUN_008808-T1 | RAN BINDING PROTEIN |
| LLU|FUN_008809-T1 | |
| LLU|FUN_008810-T1 | |
| LLU|FUN_008811-T1 | |
| LLU|FUN_008812-T1 | |
| LLU|FUN_008813-T1 | |
| LLU|FUN_008814-T1 | PROTEIN SIDEKICK |
| LLU|FUN_008815-T1 | |
| LLU|FUN_008816-T1 | NUCLEOSIDE DIPHOSPHATE KINASE 7 |
| LLU|FUN_008817-T1 | EF-HAND CALCIUM-BINDING DOMAIN CONTAINING PROTEIN |
| LLU|FUN_008818-T1 | SI:CH211-189E2.2 |
| LLU|FUN_008819-T1 | BTB/POZ DOMAIN-CONTAINING ADAPTER FOR CUL3-MEDIATED RHOA DEGRADATION PROTEIN FAMILY MEMBER |
| LLU|FUN_008820-T1 | BTB/POZ DOMAIN-CONTAINING ADAPTER FOR CUL3-MEDIATED RHOA DEGRADATION PROTEIN FAMILY MEMBER |
| LLU|FUN_008821-T1 | |
| LLU|FUN_008822-T1 | |
| LLU|FUN_008823-T1 | NUCLEOLAR GTP-BINDING PROTEIN 1 |
| LLU|FUN_008824-T1 | |
| LLU|FUN_008825-T1 | |
| LLU|FUN_008826-T1 | |
| LLU|FUN_008827-T1 | UBIQUITIN CARBOXYL-TERMINAL HYDROLASE |
| LLU|FUN_008828-T1 | SI:CH211-189E2.2 |
| LLU|FUN_008829-T1 | MOLTING PROTEIN MLT-4 |
| LLU|FUN_008830-T1 | TRANSCRIPTION INITIATION FACTOR TFIID |
| LLU|FUN_008831-T1 | CHROMOSOME-ASSOCIATED KINESIN KIF4A-RELATED |
| LLU|FUN_008832-T1 | FAT ATYPICAL CADHERIN-RELATED |

