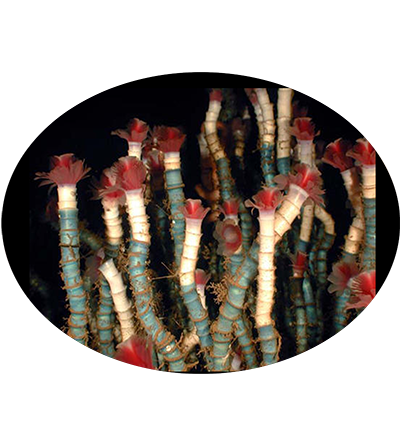
Lamellibrachia luymesi is a species of tube worms in the family Siboglinidae. It lives at deep-sea cold seeps where hydrocarbons (oil and methane) are leaking out of the seafloor. It is entirely reliant on internal, sulfide-oxidizing bacterial symbionts for its nutrition. These are located in a centrally located 'trophosome'. It provides the bacteria with hydrogen sulfide and oxygen by taking them up from the environment and binding them to a specialized hemoglobin molecule. It may help fuel the generation of sulfide by excreting sulfate through their roots into the sediments below the aggregations.
| Species | Phylum | Common Name | Ecosystem | Depth | Habitat | NCBI Taxonomy ID |
|---|---|---|---|---|---|---|
| Lamellibrachia luymesi | Annelida | cold seep-dwelling tubeworm Lamellibrachia luymesi | Hydrothermal vent | >540 | the Gulf of Mexico (N 28° 11.58′, W 89° 47.94′) | 238240 |
| Genome Assembly | Genome Size | Assembly level | Released year | WGS accession | Submitter | BioProject | BUSCO completeness | Gene Number |
|---|---|---|---|---|---|---|---|---|
| LLUY_1.0 | ~688Mb | Scaffold | 2019 | SDWI01 | Auburn University | PRJNA516467 | ~95% | 38,998 |
| Title | Journal | Pubmed ID |
|---|---|---|
| Genomic adaptations to chemosymbiosis in the deep-sea seep-dwelling tubeworm Lamellibrachia luymesi | BMC Biology | 31739792 |
| Gene ID | Description |
|---|---|
| LLU|FUN_026394-T1 | |
| LLU|FUN_026395-T1 | |
| LLU|FUN_026396-T1 | |
| LLU|FUN_026397-T1 | PDF RECEPTOR-LIKE PROTEIN-RELATED |
| LLU|FUN_026398-T1 | PDF RECEPTOR-LIKE PROTEIN-RELATED |
| LLU|FUN_026399-T1 | ZINC FINGER PROTEIN |
| LLU|FUN_026400-T1 | SPERMATOGENESIS-ASSOCIATED PROTEIN 48 |
| LLU|FUN_026401-T1 | DPY-19-LIKE 1, LIKE (H. SAPIENS) |
| LLU|FUN_026402-T1 | RING FINGER AND SWIM DOMAIN-CONTAINING PROTEIN 2 |
| LLU|FUN_026404-T1 | ARP2/3 COMPLEX 34 KDA SUBUNIT |
| LLU|FUN_026405-T1 | PROTEIN MIS12 HOMOLOG |
| LLU|FUN_026406-T1 | PROTEIN CBG20567 |
| LLU|FUN_026407-T1 | SEPIAPTERIN REDUCTASE |
| LLU|FUN_026408-T1 | 10 KDA HEAT SHOCK PROTEIN |
| LLU|FUN_026409-T1 | 60 KDA HEAT SHOCK PROTEIN, MITOCHONDRIAL |
| LLU|FUN_026410-T1 | |
| LLU|FUN_026411-T1 | POLYCYSTIN FAMILY MEMBER |
| LLU|FUN_026412-T1 | POLYCYSTIN FAMILY MEMBER |
| LLU|FUN_026413-T1 | |
| LLU|FUN_026414-T1 | |
| LLU|FUN_026415-T1 | KRUEPPEL-LIKE TRANSCRIPTION FACTOR |
| LLU|FUN_026416-T1 | |
| LLU|FUN_026417-T1 | |
| LLU|FUN_026418-T1 | ANION EXCHANGE PROTEIN |
| LLU|FUN_026419-T1 | RHOMBOID-RELATED PROTEIN |
| LLU|FUN_026420-T1 | N-ACETYLGALACTOSAMINYLTRANSFERASE |
| LLU|FUN_026421-T1 | N-ACETYLGALACTOSAMINYLTRANSFERASE |
| LLU|FUN_026422-T1 | G-PROTEIN COUPLED RECEPTOR |
| LLU|FUN_026423-T1 | |
| LLU|FUN_026424-T1 | INNEXIN |
| LLU|FUN_026425-T1 | INNEXIN |
| LLU|FUN_026426-T1 | ENOS INTERACTING PROTEIN |
| LLU|FUN_026427-T1 | MICROTUBULE ASSOCIATED PROTEIN XMAP215 |
| LLU|FUN_026428-T1 | KREV INTERACTION TRAPPED 1-RELATED |
| LLU|FUN_026429-T1 | LIPID PHOSPHATE PHOSPHATASE |
| LLU|FUN_026430-T1 | LIPID PHOSPHATE PHOSPHATASE |
| LLU|FUN_026431-T1 | |
| LLU|FUN_026432-T1 | |
| LLU|FUN_026433-T1 | LIPID PHOSPHATE PHOSPHATASE |
| LLU|FUN_026434-T1 | CALCYPHOSIN |
| LLU|FUN_026435-T1 | POLYCYSTIC KIDNEY AND HEPATIC DISEASE 1 (AUTOSOMAL RECESSIVE)-LIKE 1 |
| LLU|FUN_026436-T1 | POLYCYSTIC KIDNEY AND HEPATIC DISEASE 1 (AUTOSOMAL RECESSIVE)-LIKE 1 |
| LLU|FUN_026437-T1 | POLYCYSTIC KIDNEY AND HEPATIC DISEASE 1 (AUTOSOMAL RECESSIVE)-LIKE 1 |
| LLU|FUN_026438-T1 | |
| LLU|FUN_026439-T1 | POLYCYSTIC KIDNEY AND HEPATIC DISEASE 1 (AUTOSOMAL RECESSIVE)-LIKE 1 |
| LLU|FUN_026440-T1 | |
| LLU|FUN_026441-T1 | |
| LLU|FUN_026442-T1 | |
| LLU|FUN_026443-T1 | |
| LLU|FUN_026444-T1 |

