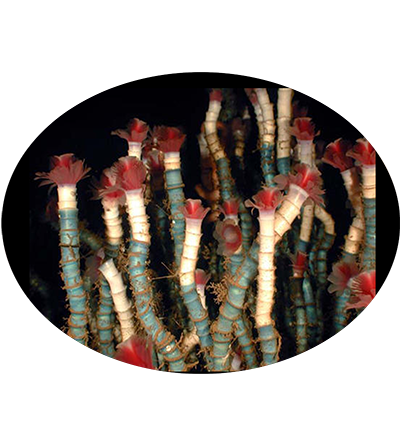
Lamellibrachia luymesi is a species of tube worms in the family Siboglinidae. It lives at deep-sea cold seeps where hydrocarbons (oil and methane) are leaking out of the seafloor. It is entirely reliant on internal, sulfide-oxidizing bacterial symbionts for its nutrition. These are located in a centrally located 'trophosome'. It provides the bacteria with hydrogen sulfide and oxygen by taking them up from the environment and binding them to a specialized hemoglobin molecule. It may help fuel the generation of sulfide by excreting sulfate through their roots into the sediments below the aggregations.
| Species | Phylum | Common Name | Ecosystem | Depth | Habitat | NCBI Taxonomy ID |
|---|---|---|---|---|---|---|
| Lamellibrachia luymesi | Annelida | cold seep-dwelling tubeworm Lamellibrachia luymesi | Hydrothermal vent | >540 | the Gulf of Mexico (N 28° 11.58′, W 89° 47.94′) | 238240 |
| Genome Assembly | Genome Size | Assembly level | Released year | WGS accession | Submitter | BioProject | BUSCO completeness | Gene Number |
|---|---|---|---|---|---|---|---|---|
| LLUY_1.0 | ~688Mb | Scaffold | 2019 | SDWI01 | Auburn University | PRJNA516467 | ~95% | 38,998 |
| Title | Journal | Pubmed ID |
|---|---|---|
| Genomic adaptations to chemosymbiosis in the deep-sea seep-dwelling tubeworm Lamellibrachia luymesi | BMC Biology | 31739792 |
| Gene ID | Description |
|---|---|
| LLU|FUN_024924-T1 | |
| LLU|FUN_024925-T1 | PAIRED BOX PROTEIN PAX-6-RELATED-RELATED |
| LLU|FUN_024926-T1 | |
| LLU|FUN_024927-T1 | |
| LLU|FUN_024928-T1 | |
| LLU|FUN_024929-T1 | |
| LLU|FUN_024930-T1 | |
| LLU|FUN_024931-T1 | |
| LLU|FUN_024932-T1 | |
| LLU|FUN_024933-T1 | |
| LLU|FUN_024934-T1 | COLLAGEN ALPHA |
| LLU|FUN_024935-T1 | EXPRESSED PROTEIN |
| LLU|FUN_024936-T1 | RGD1565685 PROTEIN |
| LLU|FUN_024937-T1 | |
| LLU|FUN_024938-T1 | |
| LLU|FUN_024939-T1 | |
| LLU|FUN_024940-T1 | |
| LLU|FUN_024941-T1 | AMYLOID PROTEIN-BINDING PROTEIN 2 |
| LLU|FUN_024942-T1 | OXYSTEROL-BINDING PROTEIN-RELATED |
| LLU|FUN_024943-T1 | AGAP001331-PA-RELATED |
| LLU|FUN_024944-T1 | |
| LLU|FUN_024945-T1 | GLYCOSYLTRANSFERASE |
| LLU|FUN_024946-T1 | |
| LLU|FUN_024947-T1 | |
| LLU|FUN_024948-T1 | |
| LLU|FUN_024949-T1 | CHYMOTRYPSIN-RELATED |
| LLU|FUN_024950-T1 | MERLIN/MOESIN/EZRIN/RADIXIN |
| LLU|FUN_024951-T1 | METALLOPHOSPHOESTERASE |
| LLU|FUN_024952-T1 | UBIQUITIN-CONJUGATING ENZYME E2 |
| LLU|FUN_024953-T1 | HFM1, ATP DEPENDENT DNA HELICASE HOMOLOG |
| LLU|FUN_024954-T1 | |
| LLU|FUN_024955-T1 | NAD-DEPENDENT PROTEIN DEACYLASE SIRTUIN-5, MITOCHONDRIAL-RELATED |
| LLU|FUN_024956-T1 | |
| LLU|FUN_024957-T1 | PDZ AND LIM DOMAIN PROTEIN ZASP |
| LLU|FUN_024958-T1 | |
| LLU|FUN_024959-T1 | PALMITOYLTRANSFERASE |
| LLU|FUN_024960-T1 | |
| LLU|FUN_024961-T1 | CALPAIN |
| LLU|FUN_024962-T1 | DENSITY-REGULATED PROTEIN HOMOLOG |
| LLU|FUN_024963-T1 | TECTONIC FAMILY MEMBER |
| LLU|FUN_024964-T1 | TRNA N6-ADENOSINE THREONYLCARBAMOYLTRANSFERASE |
| LLU|FUN_024965-T1 | TRNA N6-ADENOSINE THREONYLCARBAMOYLTRANSFERASE |
| LLU|FUN_024966-T1 | PROTEIN PHOSPHATASE 1 REGULATORY SUBUNIT 12 |
| LLU|FUN_024967-T1 | PROTEIN PHOSPHATASE 1 REGULATORY SUBUNIT 12 |
| LLU|FUN_024968-T1 | UNCHARACTERIZED |
| LLU|FUN_024969-T1 | 2,5-DIKETO-D-GLUCONIC ACID REDUCTASE |
| LLU|FUN_024970-T1 | MACROGLOBULIN / COMPLEMENT |
| LLU|FUN_024971-T1 | FMRFAMIDE RECEPTOR-RELATED |
| LLU|FUN_024972-T1 | FMRFAMIDE RECEPTOR-RELATED |
| LLU|FUN_024973-T1 | MACROGLOBULIN / COMPLEMENT |

