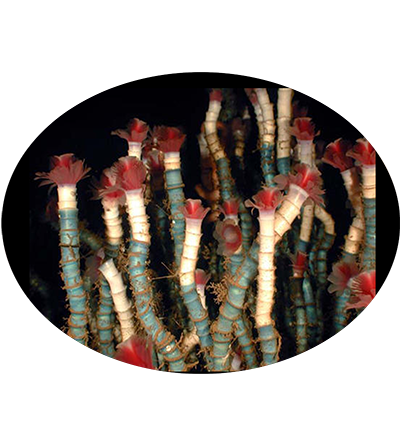
Lamellibrachia luymesi is a species of tube worms in the family Siboglinidae. It lives at deep-sea cold seeps where hydrocarbons (oil and methane) are leaking out of the seafloor. It is entirely reliant on internal, sulfide-oxidizing bacterial symbionts for its nutrition. These are located in a centrally located 'trophosome'. It provides the bacteria with hydrogen sulfide and oxygen by taking them up from the environment and binding them to a specialized hemoglobin molecule. It may help fuel the generation of sulfide by excreting sulfate through their roots into the sediments below the aggregations.
| Species | Phylum | Common Name | Ecosystem | Depth | Habitat | NCBI Taxonomy ID |
|---|---|---|---|---|---|---|
| Lamellibrachia luymesi | Annelida | cold seep-dwelling tubeworm Lamellibrachia luymesi | Hydrothermal vent | >540 | the Gulf of Mexico (N 28° 11.58′, W 89° 47.94′) | 238240 |
| Genome Assembly | Genome Size | Assembly level | Released year | WGS accession | Submitter | BioProject | BUSCO completeness | Gene Number |
|---|---|---|---|---|---|---|---|---|
| LLUY_1.0 | ~688Mb | Scaffold | 2019 | SDWI01 | Auburn University | PRJNA516467 | ~95% | 38,998 |
| Title | Journal | Pubmed ID |
|---|---|---|
| Genomic adaptations to chemosymbiosis in the deep-sea seep-dwelling tubeworm Lamellibrachia luymesi | BMC Biology | 31739792 |
| Gene ID | Description |
|---|---|
| LLU|FUN_027149-T1 | HEPARANASE |
| LLU|FUN_027150-T1 | HEPARANASE |
| LLU|FUN_027151-T1 | |
| LLU|FUN_027152-T1 | TOLL-LIKE RECEPTOR |
| LLU|FUN_027153-T1 | TOLL-LIKE RECEPTOR |
| LLU|FUN_027154-T1 | AGAP008170-PA |
| LLU|FUN_027155-T1 | SLIT RELATED LEUCINE-RICH REPEAT NEURONAL PROTEIN |
| LLU|FUN_027156-T1 | |
| LLU|FUN_027157-T1 | |
| LLU|FUN_027158-T1 | COLORECTAL MUTANT CANCER PROTEIN MCC PROTEIN -RELATED |
| LLU|FUN_027159-T1 | OS10G0105400 PROTEIN |
| LLU|FUN_027160-T1 | OS10G0105400 PROTEIN |
| LLU|FUN_027161-T1 | PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE |
| LLU|FUN_027162-T1 | |
| LLU|FUN_027163-T1 | UPF0428 PROTEIN CXORF56 |
| LLU|FUN_027164-T1 | CELL DIVISION PROTEIN KINASE |
| LLU|FUN_027165-T1 | MYCOPHENOLIC ACID ACYL-GLUCURONIDE ESTERASE, MITOCHONDRIAL |
| LLU|FUN_027166-T1 | |
| LLU|FUN_027167-T1 | IG-LIKE DOMAIN-CONTAINING PROTEIN |
| LLU|FUN_027168-T1 | HISTONE ACETYLTRANSFERASE |
| LLU|FUN_027169-T1 | HEAT SHOCK PROTEIN 70KDA |
| LLU|FUN_027170-T1 | E3 UBIQUITIN-PROTEIN LIGASE RNF26 |
| LLU|FUN_027171-T1 | |
| LLU|FUN_027172-T1 | LOW-DENSITY LIPOPROTEIN RECEPTOR-RELATED PROTEIN 2-RELATED |
| LLU|FUN_027173-T1 | LOW-DENSITY LIPOPROTEIN RECEPTOR-RELATED PROTEIN 2-RELATED |
| LLU|FUN_027174-T1 | UNCHARACTERIZED |
| LLU|FUN_027175-T1 | DIVALENT CATION TOLERANCE CUTA-RELATED |
| LLU|FUN_027176-T1 | |
| LLU|FUN_027177-T1 | HYDROXYLAMINE REDUCTASE |
| LLU|FUN_027178-T1 | |
| LLU|FUN_027179-T1 | |
| LLU|FUN_027180-T1 | MEMBRANE-SPANNING 4-DOMAINS SUBFAMILY A MS4A -RELATED |
| LLU|FUN_027181-T1 | |
| LLU|FUN_027182-T1 | |
| LLU|FUN_027183-T1 | |
| LLU|FUN_027184-T1 | CTX-RELATED TYPE I TRANSMEMBRANE PROTEIN |
| LLU|FUN_027185-T1 | |
| LLU|FUN_027186-T1 | |
| LLU|FUN_027187-T1 | TIMEOUT/TIMELESS-2 |
| LLU|FUN_027188-T1 | |
| LLU|FUN_027189-T1 | SLIT RELATED LEUCINE-RICH REPEAT NEURONAL PROTEIN |
| LLU|FUN_027190-T1 | |
| LLU|FUN_027191-T1 | |
| LLU|FUN_027192-T1 | E3 UBIQUITIN-PROTEIN LIGASE MIB2 |
| LLU|FUN_027193-T1 | SERINE/THREONINE PROTEIN KINASE |
| LLU|FUN_027194-T1 | E3 UBIQUITIN-PROTEIN LIGASE MIB2 |
| LLU|FUN_027195-T1 | FIBRINOGEN/TENASCIN/ANGIOPOEITIN |
| LLU|FUN_027196-T1 | |
| LLU|FUN_027197-T1 | |
| LLU|FUN_027198-T1 | FIBRINOGEN/TENASCIN/ANGIOPOEITIN |

