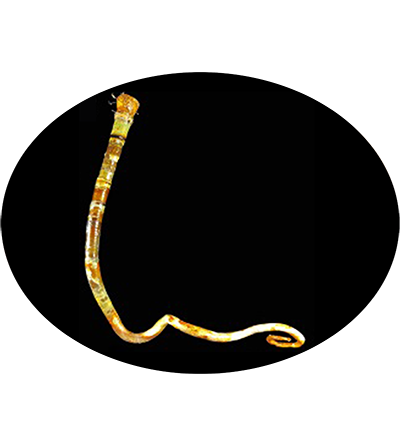
The vestimentiferan Paraescarpia echinospica is widely distributed in the cold seeps and methane seeps of the western Pacific Ocean, and relies on their endosymbiont bacteria as a source of energy and organic carbon. It's gutless and depends entirely on its endosymbiotic sulfide-oxidizing chemoautotrophic bacteria for nutrition. Its mechanisms of host–symbiont cooperation in energy production and nutrient biosynthesis and utilization have recently been documented through a study of its endosymbiont genome and metaproteome.
Animalia (Kingdom); Annelida (Phylum); Polychaeta (Class); Sedentaria (Subclass); Canalipalpata (Infraclass); Sabellida (Order); Siboglinidae (Family); Paraescarpia (Genus); Paraescarpia echinospica (Species)
Paraescarpia echinospica Southward, Schulze & Tunnicliffe, 2002
1. Southward E C, Schulze A, Tunnicliffe V. Vestimentiferans (Pogonophora) in the Pacific and Indian Oceans: a new genus from Lihir Island (Papua New Guinea) and the Java Trench, with the first report of Arcovestia ivanovi from the North Fiji Basin[J]. Journal of natural History, 2002, 36(10): 1179-1197. (Southward et al., 2010)
Edison Seamount, Lihir Island, -3.3178 (3° 19' 4" S) 152.5847 (152° 35' 5" E)
| Species | Phylum | Common Name | Ecosystem | Depth | Habitat | NCBI Taxonomy ID |
|---|---|---|---|---|---|---|
| Paraescarpia echinospica | Annelida | deep-sea tubeworm | Cold seep/Hydrothermal vent | 1100-1600 | Haima cold seep in the South China Sea (16°43.80′N, 110°28.50′E) | 2080241 |
| Genome Assembly | Genome Size | Assembly level | Released year | WGS accession | Submitter | BioProject | BUSCO completeness (%) | Scaffold/Contig N50 (kb) | GC content (%) | Repeat Rate (%) | Gene Number |
|---|---|---|---|---|---|---|---|---|---|---|---|
| HKBU_Pec_v1 | 1.09Gb | Chromosome | 2021 | JAHLWY01 | Hong Kong University of Science and Technology | PRJNA625616 | 96.40 | 67,235/253.6 | 41 | 55 | 22,642 |
| Title | Journal | Pubmed ID |
|---|---|---|
| Genomic Signatures Supporting the Symbiosis and Formation of Chitinous Tube in the Deep-Sea Tubeworm Paraescarpia echinospica | Molecular Biology and Evolution | 34255082 |
| Gene ID | Description |
|---|---|
| PE_Scaf10772_1.4 | MITOCHONDRIAL 28S RIBOSOMAL PROTEIN S28 |
| PE_Scaf10773_0.3 | NITRIC OXIDE SYNTHASE-RELATED |
| PE_Scaf10773_0.4 | COILED-COIL DOMAIN-CONTAINING PROTEIN 189 |
| PE_Scaf10773_1.0 | - |
| PE_Scaf10773_1.3 | OMEGA-6 FATTY ACID DESATURASE, CHLOROPLASTIC |
| PE_Scaf10773_2.2 | ALPHA-(1,3)-FUCOSYLTRANSFERASE C-RELATED |
| PE_Scaf10773_3.3 | EXONUCLEASE V |
| PE_Scaf10773_5.3 | - |
| PE_Scaf10773_6.11 | MYOTONIC DYSTROPHY S/T KINASE-RELATED |
| PE_Scaf10773_6.4 | - |
| PE_Scaf10774_0.4 | ENOLASE (DUF1399) |
| PE_Scaf10780_0.0 | - |
| PE_Scaf10780_0.1 | - |
| PE_Scaf10780_0.5 | EUKARYOTIC TRANSLATION INITIATION FACTOR 3 SUBUNIT G |
| PE_Scaf10780_0.6 | PROTEIN C10 |
| PE_Scaf10780_0.7 | - |
| PE_Scaf10780_1.0 | - |
| PE_Scaf10780_1.6 | SERYL-TRNA SYNTHETASE |
| PE_Scaf10780_2.4 | COMPLEMENT COMPONENT-RELATED SUSHI DOMAIN-CONTAINING |
| PE_Scaf10780_2.6 | CATION CHANNEL SPERM-ASSOCIATED PROTEIN SUBUNIT BETA |
| Annotation ID | Description | Type | Subtype | GeneRatio | Evolution Type | P-value | Q-value |
|---|---|---|---|---|---|---|---|
| GO:0006508 | proteolysis | GO | Biological process | 12 | expanded | 2.31E-07 | 2.41E-05 |
| GO:0005576 | extracellular region | GO | Cellular component | 10 | expanded | 3.02E-07 | 2.41E-05 |
| GO:0045095 | keratin filament | GO | Cellular component | 4 | expanded | 5.74E-06 | 0.000306097 |
| GO:0006352 | DNA-templated transcription, initiation | GO | Biological process | 4 | expanded | 2.55E-05 | 0.00102057 |
| GO:0043086 | negative regulation of catalytic activity | GO | Biological process | 3 | expanded | 4.91E-05 | 0.001572479 |
| GO:0008168 | methyltransferase activity | GO | Molecular function | 7 | expanded | 0.000142234 | 0.003792909 |
| GO:0032259 | methylation | GO | Biological process | 7 | expanded | 0.000219673 | 0.005021108 |
| GO:0030414 | peptidase inhibitor activity | GO | Molecular function | 3 | expanded | 0.000263266 | 0.005265319 |
| GO:0071557 | histone H3-K27 demethylation | GO | Biological process | 3 | expanded | 0.000749984 | 0.009230577 |
| GO:0071558 | histone demethylase activity (H3-K27 specific) | GO | Molecular function | 3 | expanded | 0.000749984 | 0.009230577 |
| GO:0008201 | heparin binding | GO | Molecular function | 2 | expanded | 0.0012123 | 0.01385486 |
| GO:0007399 | nervous system development | GO | Biological process | 3 | expanded | 0.001301808 | 0.013857991 |
| GO:0015031 | protein transport | GO | Biological process | 5 | expanded | 0.001547357 | 0.013857991 |
| GO:0006413 | translational initiation | GO | Biological process | 4 | expanded | 0.001818861 | 0.013857991 |
| GO:0003743 | translation initiation factor activity | GO | Molecular function | 4 | expanded | 0.001562116 | 0.013857991 |
| GO:0003723 | RNA binding | GO | Molecular function | 8 | expanded | 0.001675442 | 0.013857991 |
| GO:0005581 | collagen trimer | GO | Cellular component | 3 | expanded | 0.001495968 | 0.013857991 |
| GO:0005575 | cellular_component | GO | Cellular component | 4 | expanded | 0.0020297 | 0.014761451 |
| GO:0000272 | polysaccharide catabolic process | GO | Biological process | 2 | expanded | 0.002418597 | 0.016825021 |
| GO:0015074 | DNA integration | GO | Biological process | 3 | expanded | 0.002591176 | 0.017274507 |

