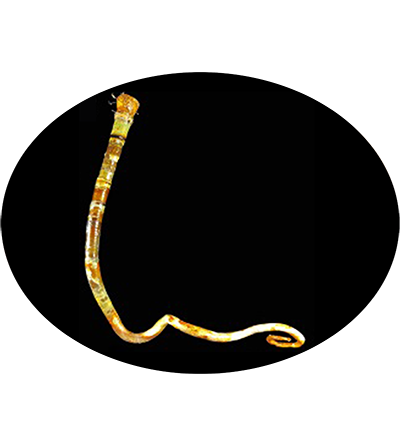
The vestimentiferan Paraescarpia echinospica is widely distributed in the cold seeps and methane seeps of the western Pacific Ocean, and relies on their endosymbiont bacteria as a source of energy and organic carbon. It's gutless and depends entirely on its endosymbiotic sulfide-oxidizing chemoautotrophic bacteria for nutrition. Its mechanisms of host–symbiont cooperation in energy production and nutrient biosynthesis and utilization have recently been documented through a study of its endosymbiont genome and metaproteome.
Animalia (Kingdom); Annelida (Phylum); Polychaeta (Class); Sedentaria (Subclass); Canalipalpata (Infraclass); Sabellida (Order); Siboglinidae (Family); Paraescarpia (Genus); Paraescarpia echinospica (Species)
Paraescarpia echinospica Southward, Schulze & Tunnicliffe, 2002
1. Southward E C, Schulze A, Tunnicliffe V. Vestimentiferans (Pogonophora) in the Pacific and Indian Oceans: a new genus from Lihir Island (Papua New Guinea) and the Java Trench, with the first report of Arcovestia ivanovi from the North Fiji Basin[J]. Journal of natural History, 2002, 36(10): 1179-1197. (Southward et al., 2010)
Edison Seamount, Lihir Island, -3.3178 (3° 19' 4" S) 152.5847 (152° 35' 5" E)
| Species | Phylum | Common Name | Ecosystem | Depth | Habitat | NCBI Taxonomy ID |
|---|---|---|---|---|---|---|
| Paraescarpia echinospica | Annelida | deep-sea tubeworm | Cold seep/Hydrothermal vent | 1100-1600 | Haima cold seep in the South China Sea (16°43.80′N, 110°28.50′E) | 2080241 |
| Genome Assembly | Genome Size | Assembly level | Released year | WGS accession | Submitter | BioProject | BUSCO completeness (%) | Scaffold/Contig N50 (kb) | GC content (%) | Repeat Rate (%) | Gene Number |
|---|---|---|---|---|---|---|---|---|---|---|---|
| HKBU_Pec_v1 | 1.09Gb | Chromosome | 2021 | JAHLWY01 | Hong Kong University of Science and Technology | PRJNA625616 | 96.40 | 67,235/253.6 | 41 | 55 | 22,642 |
| Title | Journal | Pubmed ID |
|---|---|---|
| Genomic Signatures Supporting the Symbiosis and Formation of Chitinous Tube in the Deep-Sea Tubeworm Paraescarpia echinospica | Molecular Biology and Evolution | 34255082 |
| Gene ID | Description |
|---|---|
| PE_Scaf10000_2.0 | - |
| PE_Scaf10000_3.0 | - |
| PE_Scaf10000_3.5 | METHANETHIOL OXIDASE |
| PE_Scaf10000_4.5 | - |
| PE_Scaf10000_6.0 | - |
| PE_Scaf10000_6.7 | S-ADENOSYL-L-METHIONINE:CARBOXYL METHYLTRANSFERASE FAMILY PROTEIN |
| PE_Scaf10000_6.8 | OVOCHYMASE-RELATED |
| PE_Scaf10001_0.1 | - |
| PE_Scaf10001_0.4 | - |
| PE_Scaf10002_1.0 | GALACTOSE-3-O-SULFOTRANSFERASE |
| PE_Scaf10002_4.10 | UNC-13-4A, ISOFORM B |
| PE_Scaf10002_4.11 | EXTRACELLULAR MATRIX GLYCOPROTEIN RELATED |
| PE_Scaf10002_4.5 | ATP-DEPENDENT DNA HELICASE |
| PE_Scaf10002_5.10 | - |
| PE_Scaf10002_5.15 | ATP-DEPENDENT DNA HELICASE |
| PE_Scaf10002_5.17 | - |
| PE_Scaf10002_5.2 | - |
| PE_Scaf10003_0.10 | DBX |
| PE_Scaf10003_0.14 | - |
| PE_Scaf10003_0.9 | ZGC:172139 |
| Annotation ID | Description | Type | Subtype | GeneRatio | Evolution Type | P-value | Q-value |
|---|---|---|---|---|---|---|---|
| GO:0010468 | regulation of gene expression | GO | Biological process | 3 | expanded | 0.002736508 | 0.017513649 |
| GO:0005604 | basement membrane | GO | Cellular component | 2 | expanded | 0.003167879 | 0.019494638 |
| GO:0006030 | chitin metabolic process | GO | Biological process | 3 | expanded | 0.003895322 | 0.02308339 |
| GO:0008233 | peptidase activity | GO | Molecular function | 5 | expanded | 0.004602371 | 0.026299262 |
| GO:0008061 | chitin binding | GO | Molecular function | 3 | expanded | 0.004881228 | 0.026930915 |
| GO:0008234 | cysteine-type peptidase activity | GO | Molecular function | 3 | expanded | 0.005094833 | 0.027172444 |
| GO:0030198 | extracellular matrix organization | GO | Biological process | 2 | expanded | 0.005451028 | 0.027255139 |
| GO:0044666 | MLL3/4 complex | GO | Cellular component | 2 | expanded | 0.005451028 | 0.027255139 |
| GO:0031490 | chromatin DNA binding | GO | Molecular function | 2 | expanded | 0.00597595 | 0.028974303 |

