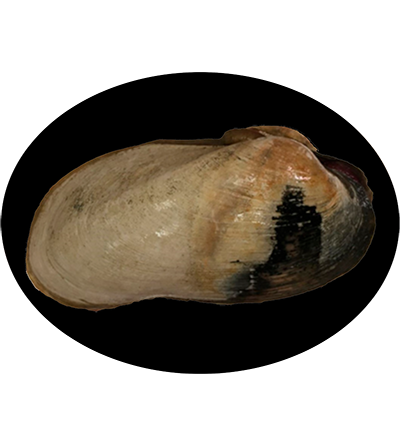
The Deep-Sea Chemosymbiotic Clam Archivesica marissinica is a species of deep-sea clam in the family Vesicomyidae, commonly found at the Haima cold seep on the northwest continental slope of the South China Sea. This species is well-adapted to extreme environments and relies on symbiotic sulfur-oxidizing bacteria housed in its gills for nutrition. Archivesica marissinica plays a significant role in cold seep ecosystems by contributing to nutrient cycling and supporting diverse microbial and faunal communities.
Animalia (Kingdom); Mollusca (Phylum); Bivalvia (Class); Autobranchia (Subclass); Venerida (Order); Vesicomyidae (Family); Archivesica (Genus); Archivesica marissinica (Species)
Archivesica marissinica (Chen, Okutani, Liang & Qiu, 2018)
1. Chen C, Okutani T, Liang Q, et al. A noteworthy new species of the family Vesicomyidae from the South China Sea (Bivalvia: Glossoidea)[J]. Venus (Journal of the Malacological Society of Japan), 2018, 76(1-4): 29-37. (Chen et al., 2018)
Shell thin, rather lightly-built, chalky, elongate-oval in profile, slightly attenuated in front and somewhat flared posteriorly. Shell height about a quarter of shell length. Umbo situated at about 20% anteriorly, with weakly prosogyrous beaks. External surface covered by straw-colored, varnished periostracum, which is occasionally commarginally lamellate, particularly in front of umbonal region and marginal region. Periostracum corroded behind umbonal region suggesting presence of boundary between buried portion and exposed portion. Round antero- dorsal and anterior margins gently continuous to sub-straight ventral margin. No lunule present. Posterior margin also round. A low, obscure radial ridge running from umbo to postero-ventral corner. Escutcheon not demarcated, dorso-posterior margin weakly convex, representing greatest position in shell height, and generating nearly flat area between dorsal margin and dorsal rim of radial ridge. Ligament amphidetic, strong, extending about half length from umbo to posterior tip. Calcareous fibrous layer rather weak
meaning “Chinese Sea”
Hitherto known only from the type locality: Haima methane seep, south off Hainan Island. 1,372–1,398 meters depth
| Species | Phylum | Common Name | Ecosystem | Depth | Habitat | NCBI Taxonomy ID |
|---|---|---|---|---|---|---|
| Archivesica marissinica | Mollusca | Deep-Sea Clam | Hydrothermal vent | 1,361 | Haima cold seep in the South China Sea | 2291877 |
| Genome Assembly | Genome Size | Assembly level | Released year | WGS accession | Submitter | BioProject | BUSCO completeness (%) | Scaffold/Contig N50 (kb) | GC content (%) | Repeat Rate (%) | Gene Number |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ASM1484369v1 | 1.5Gb | Chromosome | 2020 | JABXNX01 | Hong Kong Baptist University | PRJNA629898 | 93.50 | 74,300/79.1 | 39 | 52.81 | 28,949 |
| Title | Journal | Pubmed ID |
|---|---|---|
| Host-Endosymbiont Genome Integration in a Deep-Sea Chemosymbiotic Clam | Molecular Biology and Evolution | 32956455 |
| Gene ID | Description |
|---|---|
| Ama27973 | - |
| Ama27976 | 52 KDA REPRESSOR OF THE INHIBITOR OF THE PROTEIN KINASE-LIKE PROTEIN-RELATED |
| Ama27978 | CILIA- AND FLAGELLA-ASSOCIATED PROTEIN 99 |
| Ama27980 | G PROTEIN-COUPLED RECEPTOR KINASE/RIBOSOMAL PROTEIN S6 KINASE |
| Ama27980.1.5db7de70 | G PROTEIN-COUPLED RECEPTOR KINASE/RIBOSOMAL PROTEIN S6 KINASE |
| Ama27981 | CILIA AND FLAGELLA-ASSOCIATED PROTEIN 58-RELATED |
| Ama27982 | COMPLEMENT COMPONENT-RELATED SUSHI DOMAIN-CONTAINING |
| Ama27983 | - |
| Ama27985 | SOLUTE CARRIER FAMILY 17 |
| Ama27986 | SOLUTE CARRIER FAMILY 17 |
| Ama27987 | SOLUTE CARRIER FAMILY 17 |
| Ama27988 | SOLUTE CARRIER FAMILY 17 |
| Ama27989 | SOLUTE CARRIER FAMILY 17 |
| Ama27990 | FUCOSE MUTAROTASE |
| Ama27991 | TRANSFORMING GROWTH FACTOR BETA REGULATED GENE 1 |
| Ama27992 | GLUCOSE-METHANOL-CHOLINE GMC OXIDOREDUCTASE |
| Ama27993 | - |
| Ama27996 | THAP DOMAIN PROTEIN |
| Ama27999 | POLYCYSTIN FAMILY MEMBER |
| Ama27_5db7b01a | - |
| Annotation ID | Description | Type | Subtype | GeneRatio | Evolution Type | P-value | Q-value |
|---|---|---|---|---|---|---|---|
| GO:0035652 | cargo loading into clathrin-coated vesicle | GO | Biological Process | 47 | expanded | - | 6.98E-08 |
| GO:0060260 | regulation of transcription initiation from RNA polymerase II promoter | GO | Biological Process | 92 | expanded | - | 1.96E-05 |
| GO:0035459 | cargo loading into vesicle | GO | Biological Process | 61 | expanded | - | 2.24E-05 |
| GO:0043939 | negative regulation of sporulation | GO | Biological Process | 22 | expanded | - | 0.000380302 |
| GO:0007062 | sister chromatid cohesion | GO | Biological Process | 83 | expanded | - | 0.00058477 |
| GO:0033363 | secretory granule organization | GO | Biological Process | 85 | expanded | - | 0.001045836 |
| GO:0043937 | regulation of sporulation | GO | Biological Process | 22 | expanded | - | 0.001181872 |
| GO:0034307 | regulation of ascospore formation | GO | Biological Process | 22 | expanded | - | 0.001242481 |
| GO:0071444 | cellular response to pheromone | GO | Biological Process | 22 | expanded | - | 0.003797717 |
| GO:0006384 | transcription initiation from RNA polymerase III promoter | GO | Biological Process | 30 | expanded | - | 0.005224062 |
| GO:0019236 | response to pheromone | GO | Biological Process | 24 | expanded | - | 0.009234132 |
| GO:0048918 | posterior lateral line nerve development | GO | Biological Process | 19 | expanded | - | 0.019616942 |
| GO:0006302 | double-strand break repair | GO | Biological Process | 227 | expanded | - | 0.019943185 |
| GO:0090083 | regulation of inclusion body assembly | GO | Biological Process | 48 | expanded | - | 0.023871281 |
| GO:0060876 | semicircular canal formation | GO | Biological Process | 18 | expanded | - | 0.027381017 |
| GO:0032290 | peripheral nervous system myelin formation | GO | Biological Process | 18 | expanded | - | 0.027710909 |
| GO:0070230 | positive regulation of lymphocyte apoptotic process | GO | Biological Process | 55 | expanded | - | 0.037668635 |
| GO:0009617 | response to bacterium | GO | Biological Process | 374 | contracted | - | 0.009719681 |
| GO:0034622 | cellular macromolecular complex assembly | GO | Biological Process | 674 | contracted | - | 0.01744258 |
| GO:0051248 | negative regulation of protein metabolic process | GO | Biological Process | 672 | contracted | - | 0.028576899 |

