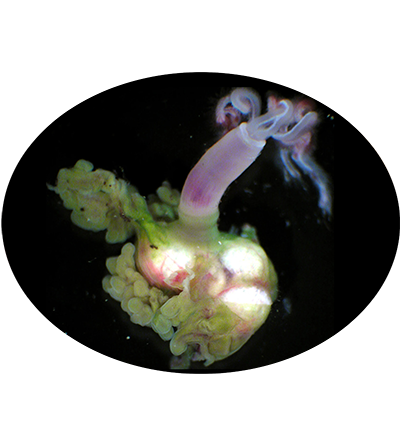
Osedax frankpressi is a species of bathypelagic polychaete worm that lives on the seabed. It is found on the carcase of a gray whale in Monterey Canyon, off the coast of California, at a depth of 2,891 m. And it is a small worm that speedily colonises the skeleton, sending out root-like threads that force their way into the bone marrow and absorb the nutrients.[5] Inside these root structures are bacteria in the order Oceanospirillales with which the worm is in symbiosis. It is probable that the presence of these bacteria, with their ability to metabolise organic material, enables the worms to live on carcases.
Animalia (Kingdom); Annelida (Phylum); Polychaeta (Class); Sedentaria (Subclass); Canalipalpata (Infraclass); Sabellida (Order); Siboglinidae (Family); Osedax (Genus); Osedax frankpressi (Species)
Osedax frankpressi Rouse, Goffredi & Vrijenhoek, 2004
1. Rouse G W, Goffredi S K, Vrijenhoek R C. Osedax: bone-eating marine worms with dwarf males[J]. Science, 2004, 305(5684): 668-671. (Rouse et al., 2004)
Holotype, emergent body in gelatinous hemispherical tube, 7-mm diameter. Contracted crown plumes 0.95 cm long. Oviduct filled with ellipsoid eggs, Oviduct convoluted upon contraction, extending between palps, 3 mm from trunk. Palps red with two longitudinal white stripes in living worms; pinnules on inner margins. Trunk 4.5 mm long, 0.9 mm wide, and marked by white thickened tissue at anterior. Green, bacteriocyte-filled sheath forms trunk-ovisac junction 1.2 mm long, 1 mm wide. Lobulate ovisac, 6.5 mm by 5 mm by 3 mm. Ovisac and roots inflated with clear fluid in situ in bone; fluid lost on extraction. Allotypes, 0.15- to 0.25-mm-long males. Chaetae with hooks and handles 15 to 21 μm. Capitium with five teeth; no subrostral teeth.
In honor of Dr. Frank Press, former U.S. presidential science advisor, president of the U.S. National Academy of Sciences, and chair of the MBARI Board of Directors, for his distinguished service to science
Monterey Bay, California, Tiburon dive T610, 36°36.8'N, 122°26.0'W, 2891 m
| Species | Phylum | Common Name | Ecosystem | Depth | Habitat | NCBI Taxonomy ID |
|---|---|---|---|---|---|---|
| Osedax frankpressi | Annelida | deep-sea annelid worms | Hydrothermal vent | 1018 or 2,891 | coasts of California and Mexico | 283776 |
| Genome Assembly | Genome Size | Assembly level | Released year | WGS accession | Submitter | BioProject | BUSCO completeness (%) | Scaffold/Contig N50 (kb) | GC content (%) | Repeat Rate (%) | Gene Number |
|---|---|---|---|---|---|---|---|---|---|---|---|
| - | 285Mb | Scaffold | 2023 | - | HARVARD UNIVERSITY | PRJNA283630 | 80.10 | 426 | 29.08 | 29.16 | 14,203 |
| Title | Journal | Pubmed ID |
|---|---|---|
| Distinct genomic routes underlie transitions to specialised symbiotic lifestyles in deep-sea annelid worms | nature communications | 37198188 |
| Gene ID | Description |
|---|---|
| OFRAG00000000138.1 | - |
| OFRAG00000000139.1 | - |
| OFRAG00000000140.1 | MATRIX METALLOPROTEINASE |
| OFRAG00000000141.1 | - |
| OFRAG00000000142.1 | - |
| OFRAG00000000143.1 | - |
| OFRAG00000000144.1 | ZINC METALLOPROTEASE FAMILY M13 NEPRILYSIN-RELATED |
| OFRAG00000000145.1 | ZINC METALLOPROTEASE FAMILY M13 NEPRILYSIN-RELATED |
| OFRAG00000000146.1 | - |
| OFRAG00000000147.1 | ZINC METALLOPROTEASE FAMILY M13 NEPRILYSIN-RELATED |
| OFRAG00000000148.1 | - |
| OFRAG00000000149.1 | ZINC METALLOPROTEASE FAMILY M13 NEPRILYSIN-RELATED |
| OFRAG00000000150.1 | - |
| OFRAG00000000151.1 | ZINC METALLOPROTEASE FAMILY M13 NEPRILYSIN-RELATED |
| OFRAG00000000152.1 | - |
| OFRAG00000000153.1 | MITOCHONDRIAL METAL TRANSPORTER 1-RELATED |
| OFRAG00000000154.1 | MITOCHONDRIAL METAL TRANSPORTER 1-RELATED |
| OFRAG00000000155.1 | MYOSIN LIGHT CHAIN 1, 3 |
| OFRAG00000000156.1 | ARRESTIN DOMAIN CONTAINING PROTEIN |
| OFRAG00000000157.1 | REGULATOR OF CHROMOSOME CONDENSATION |

