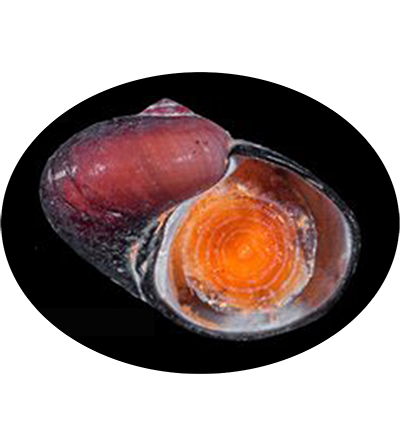
Dracogyra subfuscus is a species of deep-sea gastropod in the family Peltospiridae, currently known only from the Longqi hydrothermal vent field on the Southwest Indian Ridge. This small gastropod is adapted to extreme environments, thriving in the high-temperature and low-oxygen conditions of hydrothermal vents. Its discovery highlights the unique biodiversity of vent ecosystems and provides valuable insights into the adaptations of organisms to these harsh habitats.
Animalia (Kingdom); Mollusca (Phylum); Gastropoda (Class); Vetigastropoda (Subclass); Neomphalida (Order); Neomphaloidea (Superfamily); Peltospiridae (Family); Dracogyra (Genus); Dracogyra subfuscus (Species)
Dracogyra subfuscus C. Chen, Y.-D. Zhou, C.-S. Wang & Copley, 2017
1. Chen C, Zhou Y, Wang C, et al. Two new hot-vent peltospirid snails (Gastropoda: Neomphalina) from Longqi hydrothermal field, Southwest Indian Ridge[J]. Frontiers in Marine Science, 2017, 4: 392. (Chen et al., 2017)
Shell tightly coiled, strongly depressed with a low spire, shape between skeneiform and neritiform. Medium-sized for the family, the largest specimen available (a dead shell) measured 4.6 mm in shell height and 6.3 mm in shell width. More measurements shown in Table 2. Protoconch indistinctly coiled, 200 μm in diameter. Specimen with protoconch sculpture preserved was not found in materials available. Teleoconch white and lacks nacreous layer, moderately thick, consist of about 2.7 whorls, depressed oval in cross section. Perfectly smooth except fine growth striations. Aperture opisthocline to the coiling axis, oval in shape, lacking any significant notch. Outer lip simple, not thickened in adults. Inner lip with a small section of columellar callus extending toward the umbilical area. Umbilicus extremely narrow, sometimes obscured by callus. Microstructure of a thin, granular outer layer and a thick, cross-lamellar inner layer. The inner layer bears shell pores which open to the interior of the teleoconch, pores do not reach the outer layer. Periostracum thick, leathery. Initially light reddish brown, gradually darkening to near-black in last 0.5 whorls of adults. Lacks significant sculpture or pattern except fine growth lines. Outer edge extends over the outer lip, enveloping it. Never seen covered by thick mineral deposits in specimens available, although scattered deposits and evidence of corrosion is commonplace.
“Draco” (Latin), dragon; “gyrus” (Latin), to circle or coil. Named for the Longqi vent field; “Longqi” means “dragon flag” in Chinese and “Dragon vent field” has been used as an alternative name
Longqi vent field, Southwest Indian Ridge, 37°47.03′ S, 49°38.97′E (“Tiamat” Chimney), 2785 m deep, RRS James Cook expedition JC67, ROV Kiel6000 Dive 142
| Species | Phylum | Common Name | Ecosystem | Depth | Habitat | NCBI Taxonomy ID |
|---|---|---|---|---|---|---|
| Dracogyra subfusca | Mollusca | Dracogyra subfuscus | Hydrothermal vent | 2,736-2,785 | Longqi (‘Tiamat’ chimney) | 2038759 |
| Genome Assembly | Genome Size | Assembly level | Released year | WGS accession | Submitter | BioProject | BUSCO completeness (%) | Scaffold/Contig N50 (kb) | GC content (%) | Repeat Rate (%) | Gene Number |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ASM1610662v1 | 1.16Gb | Scaffold | 2020 | JAECMU01 | Hong Kong University of Science and Technology | PRJNA680542 | 32.70 | 5.9/4.1 | 37 | 56 | 5,986 |
| Title | Journal | Pubmed ID |
|---|---|---|
| Hologenome analysis reveals dual symbiosis in the deep-sea hydrothermal vent snail Gigantopelta aegis | nature communications | 33608555 |
| Gene ID | Description |
|---|---|
| scaffold133081_cov125_0.0 | WHEY ACDIC PROTEIN WAP |
| scaffold133091_cov157_0.0 | MITOCHONDRIAL DIVISION PROTEIN 1-RELATED |
| scaffold133105_cov108_0.0 | ZINC FINGER PROTEIN |
| scaffold133221_cov141_0.0 | ZINC FINGER PROTEIN |
| scaffold133373_cov144_0.1 | HEAT SHOCK PROTEIN 70KDA |
| scaffold133451_cov133_0.1 | SET AND MYND DOMAIN-CONTAINING PROTEIN 5 |
| scaffold13353_cov142_0.0 | UNCHARACTERIZED |
| scaffold133603_cov171_0.1 | - |
| scaffold133703_cov90_0.0 | HOMEO BOX HB9 LIKE A-RELATED |
| scaffold133834_cov114_0.0 | NEGATIVE ELONGATION FACTOR D |
| scaffold133892_cov149_0.0 | - |
| scaffold133918_cov156_0.0 | - |
| scaffold133986_cov144_0.0 | - |
| scaffold134020_cov150_0.0 | NUCLEAR HORMONE RECEPTOR |
| scaffold134020_cov150_0.2 | - |
| scaffold134035_cov112_0.0 | TUMOR NECROSIS FACTOR FAMILY MEMBER |
| scaffold134121_cov122_0.0 | - |
| scaffold134178_cov141_0.0 | - |
| scaffold13423_cov178_0.0 | - |
| scaffold134255_cov176_0.0 | PROTEIN HGH1 HOMOLOG |

